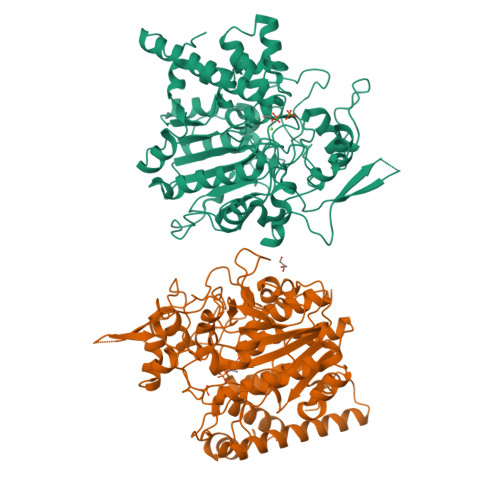
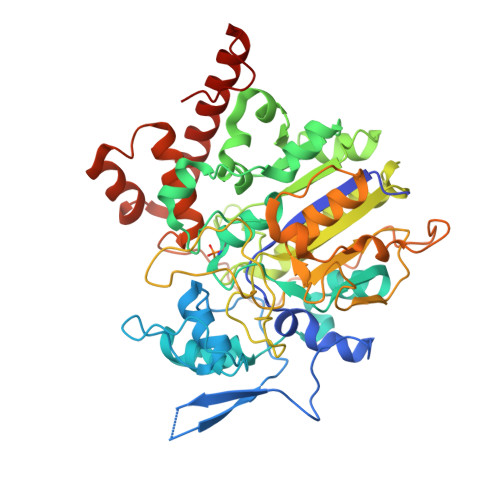
Insights into the mechanism of phospholipid hydrolysis by plant non-specific phospholipase C.
Fan, R., Zhao, F., Gong, Z., Chen, Y., Yang, B., Zhou, C., Zhang, J., Du, Z., Wang, X., Yin, P., Guo, L., Liu, Z.(2023) Nat Commun 14: 194-194
- PubMed: 36635324
- DOI: https://doi.org/10.1038/s41467-023-35915-4
- Primary Citation of Related Structures:
8HAV, 8HAW - PubMed Abstract:
Non-specific phospholipase C (NPC) hydrolyzes major membrane phospholipids to release diacylglycerol (DAG), a potent lipid-derived messenger regulating cell functions. Despite extensive studies on NPCs reveal their fundamental roles in plant growth and development, the mechanistic understanding of phospholipid-hydrolyzing by NPCs, remains largely unknown. Here we report the crystal structure of Arabidopsis NPC4 at a resolution of 2.1 Å. NPC4 is divided into a phosphoesterase domain (PD) and a C-terminal domain (CTD), and is structurally distinct from other characterized phospholipases. The previously uncharacterized CTD is indispensable for the full activity of NPC4. Mechanistically, CTD contributes NPC4 activity mainly via CTD α1 -PD interaction, which ultimately stabilizes the catalytic pocket in PD. Together with a series of structure-guided biochemical studies, our work elucidates the structural basis and provides molecular mechanism of phospholipid hydrolysis by NPC4, and adds new insights into the members of phospholipase family.
Organizational Affiliation:
National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, 430070, China.