Discovery, SAR Study of GST Inhibitors from a Novel Quinazolin-4(1 H )-one Focused DNA-Encoded Library.
Wen, X., Zhang, M., Duan, Z., Suo, Y., Lu, W., Jin, R., Mu, B., Li, K., Zhang, X., Meng, L., Hong, Y., Wang, X., Hu, H., Zhu, J., Song, W., Shen, A., Lu, X.(2023) J Med Chem 66: 11118-11132
- PubMed: 37552553
- DOI: https://doi.org/10.1021/acs.jmedchem.2c02129
- Primary Citation of Related Structures:
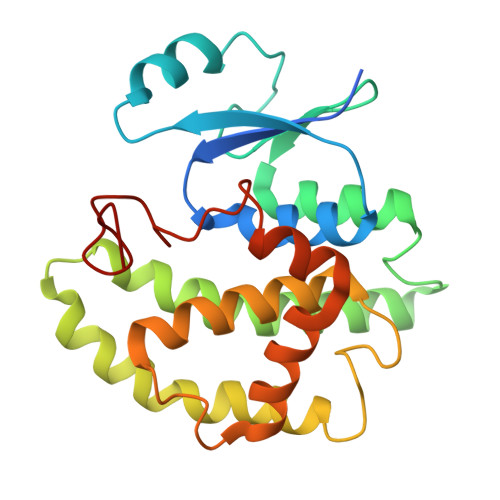
8GYD - PubMed Abstract:
The DNA-encoded library (DEL) is a powerful hit-generation tool in drug discovery. This study describes a new DEL with a privileged scaffold quinazolin-4(3 H )-one developed by a robust DNA-compatible multicomponent reaction and a series of novel glutathione S -transferase (GST) inhibitors that were identified through affinity-mediated DEL selection. A novel inhibitor 16 was subsequently verified with an inhibitory potency value of 1.55 ± 0.02 μM against SjGST and 2.02 ± 0.20 μM against hGSTM2. Further optimization was carried out via various structure-activity relationship studies. And especially, the co-crystal structure of the compound 16 with the SjGST was unveiled, which clearly demonstrated its binding mode was quite different from the known GSH-like compounds. This new type of probe is likely to play a different role compared with the GSH, which may provide new opportunities to discover more potent GST inhibitors.
- State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, 501 Haike Road, Zhang Jiang Hi-Tech Park, Pudong, Shanghai 201203, P. R. China.
Organizational Affiliation: