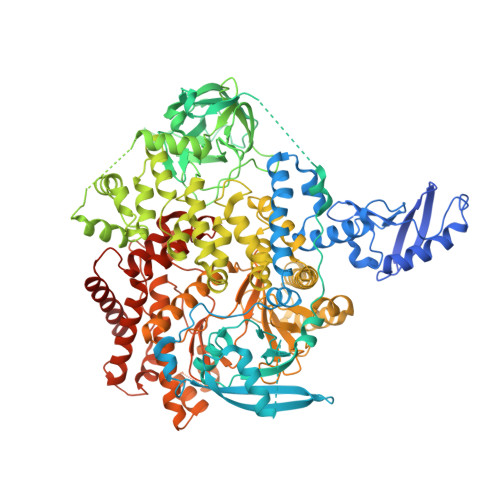
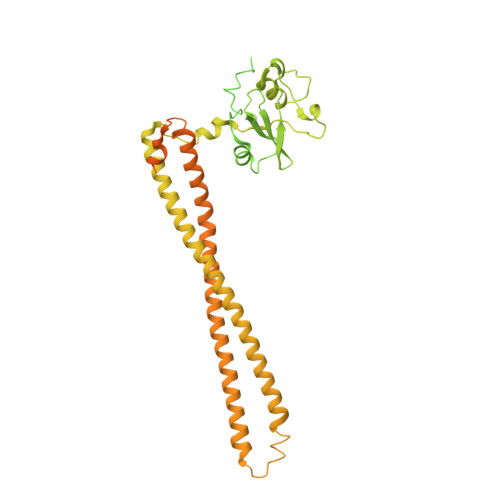
Cryo-EM structures of cancer-specific helical and kinase domain mutations of PI3K alpha.
Liu, X., Zhou, Q., Hart, J.R., Xu, Y., Yang, S., Yang, D., Vogt, P.K., Wang, M.W.(2022) Proc Natl Acad Sci U S A 119: e2215621119-e2215621119
- PubMed: 36343266
- DOI: https://doi.org/10.1073/pnas.2215621119
- Primary Citation of Related Structures:
8GUA, 8GUB, 8GUD - PubMed Abstract:
Phosphoinositide 3-kinases (PI3Ks) are a family of lipid kinases that perform multiple and important cellular functions. The protein investigated here belongs to class IA of the PI3Ks; it is a dimer consisting of a catalytic subunit, p110α, and a regulatory subunit, p85α, and is referred to as PI3Kα. The catalytic subunit p110α is frequently mutated in cancer. The mutations induce a gain of function and constitute a driving force in cancer development. About 80% of these mutations lead to single-amino-acid substitutions in one of three sites of p110α: two in the helical domain of the protein (E542K and E545K) and one at the C-terminus of the kinase domain (H1047R). Here, we report the cryo-electron microscopy structures of these mutants in complex with the p110α-specific inhibitor BYL-719. The H1047R mutant rotates its sidechain to a new position and weakens the kα11 activation loop interaction, thereby reducing the inhibitory effect of p85α on p110α. E542K and E545K completely abolish the tight interaction between the helical domain of p110α and the N-terminal SH2 domain of p85α and lead to the disruption of all p85α binding and a dramatic increase in flexibility of the adaptor-binding domain (ABD) in p110α. Yet, the dimerization of PI3Kα is preserved through the ABD-p85α interaction. The local and global structural features induced by these mutations provide molecular insights into the activation of PI3Kα, deepen our understanding of the oncogenic mechanism of this important signaling molecule, and may facilitate the development of mutant-specific inhibitors.
- Department of Pharmacology, School of Basic Medical Sciences, Fudan University, Shanghai 200032, China.
Organizational Affiliation: