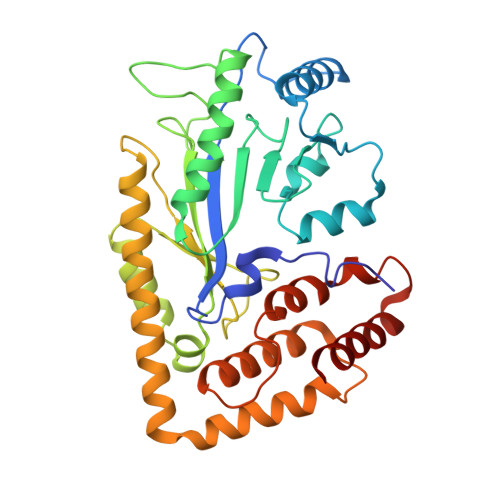
Complex structure and activation mechanism of arginine kinase McsB by McsA.
Lu, K., Luo, B., Tao, X., Luo, Y., Ao, M., Zheng, B., Xu, X., Ma, X., Niu, J., Li, H., Xie, Y., Zhao, Z., Zheng, P., Wang, G., Gao, S., Wang, C., Xia, W., Su, Z., Mao, Z.W.(2024) Nat Chem Biol
- PubMed: 39232187
- DOI: https://doi.org/10.1038/s41589-024-01720-3
- Primary Citation of Related Structures:
8GQD - PubMed Abstract:
Protein phosphorylation is a pivotal post-translational modification modulating various cellular processes. In Gram-positive bacteria, the protein arginine kinase McsB, along with its activator McsA, has a key role in labeling misfolded and damaged proteins during stress. However, the activation mechanism of McsB by McsA remains elusive. Here we report the cryo-electron microscopy structure of a tetrameric McsA-McsB complex at 3.41 Å resolution. Biochemical analysis indicates that the homotetrameric assembly is essential for McsB's kinase activity. The conserved C-terminal zinc finger of McsA interacts with an extended loop in McsB, optimally orienting a critical catalytic cysteine residue. In addition, McsA binding decreases the CtsR's affinity for McsB, enhancing McsB's kinase activity and accelerating the turnover rate of CtsR phosphorylation. Furthermore, McsA binding also increases McsB's thermostability, ensuring its activity under heat stress. These findings elucidate the structural basis and activation mechanism of McsB in stress response.
- MOE Key Laboratory of Bioinorganic and Synthetic Chemistry, School of Chemistry, IGCME, GBRCE for Functional Molecular Engineering, Sun Yat-Sen University, Guangzhou, China.
Organizational Affiliation: