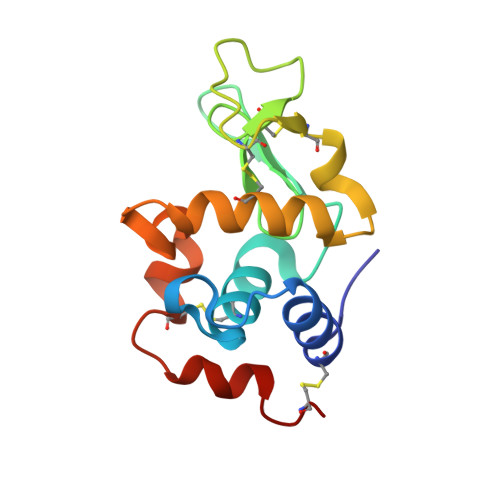
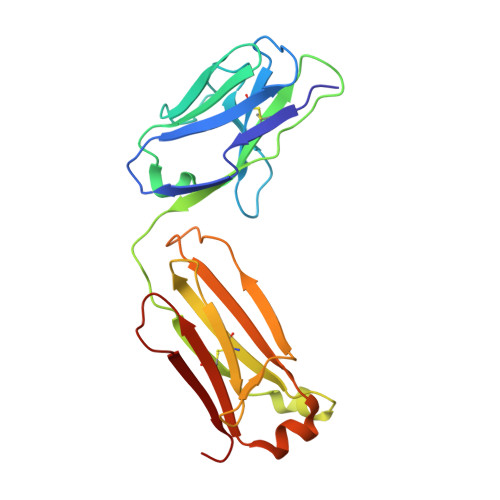
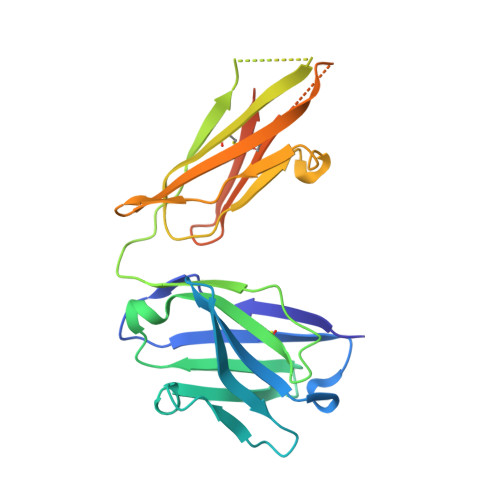
Arginine cluster introduction on framework region in anti-lysozyme antibody improved association rate constant by changing conformational diversity of CDR loops.
Maeta, S., Nakakido, M., Matsuura, H., Sakai, N., Hirata, K., Kuroda, D., Fukunaga, A., Tsumoto, K.(2023) Protein Sci 32: e4745-e4745
- PubMed: 37550885
- DOI: https://doi.org/10.1002/pro.4745
- Primary Citation of Related Structures:
8GQ1 - PubMed Abstract:
Antibodies are used for many therapeutic and biotechnological purposes. Because the affinity of an antibody to the antigen is critical for clinical efficacy of pharmaceuticals, many affinity maturation strategies have been developed. Although we previously reported an affinity maturation strategy in which the association rate of the antibody toward its antigen is improved by introducing a cluster of arginine residues into the framework region of the antibody, the detailed molecular mechanism responsible for this improvement has been unknown. In this study, we introduced five arginine residues into an anti-hen egg white lysozyme antibody (HyHEL10) Fab fragment to create the R5-mutant and comprehensively characterized the interaction between antibody and antigen using thermodynamic analysis, X-ray crystallography, and molecular dynamics (MD) simulations. Our results indicate that introduction of charged residues strongly enhanced the association rate, as previously reported, and the antibody-antigen complex structure was almost the same for the R5-mutant and wild-type Fabs. The MD simulations indicate that the mutation increased conformational diversity in complementarity-determining region loops and thereby enhanced the association rate. These observations provide the molecular basis of affinity maturation by R5 mutation.
Organizational Affiliation:
Bio-Diagnostic Reagent Technology Center, Sysmex Corporation, Kobe, Japan.