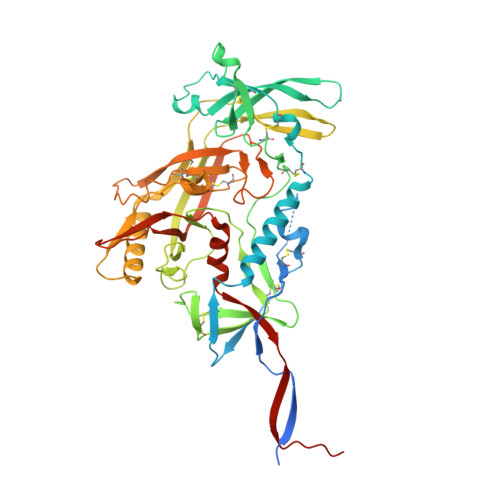
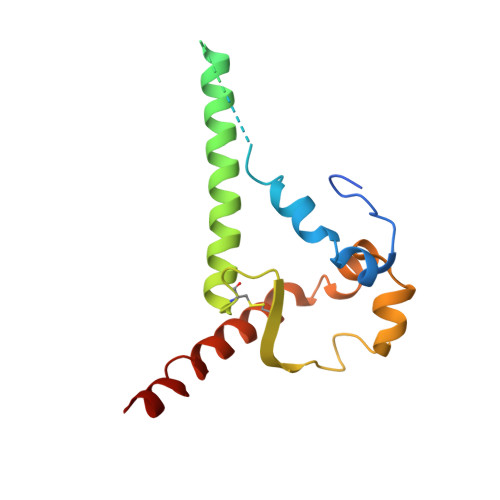
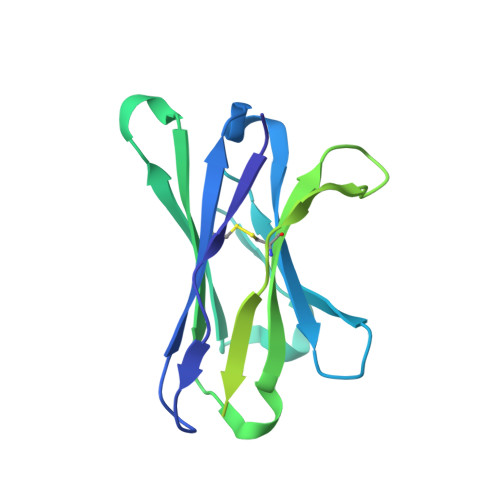
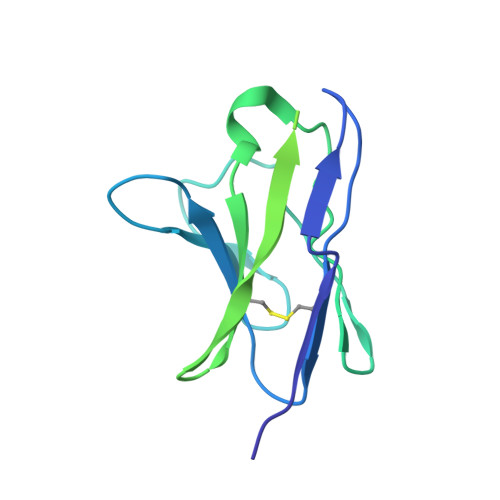
Glycan heterogeneity as a cause of the persistent fraction in HIV-1 neutralization.
Ringe, R.P., Colin, P., Ozorowski, G., Allen, J.D., Yasmeen, A., Seabright, G.E., Lee, J.H., Antanasijevic, A., Rantalainen, K., Ketas, T., Moore, J.P., Ward, A.B., Crispin, M., Klasse, P.J.(2023) PLoS Pathog 19: e1011601-e1011601
- PubMed: 37903160
- DOI: https://doi.org/10.1371/journal.ppat.1011601
- Primary Citation of Related Structures:
8GJE - PubMed Abstract:
Neutralizing antibodies (NAbs) to multiple epitopes on the HIV-1-envelope glycoprotein (Env) have been isolated from infected persons. The potency of NAbs is measured more often than the size of the persistent fraction of infectivity at maximum neutralization, which may also influence preventive efficacy of active or passive immunization and the therapeutic outcome of the latter. Many NAbs neutralize HIV-1 CZA97.012, a clone of a Clade-C isolate, to ~100%. But here NAb PGT151, directed to a fusion-peptide epitope, left a persistent fraction of 15%. NAb PGT145, ligating the Env-trimer apex, left no detectable persistent fraction. The divergence in persistent fractions was further analyzed by depletion of pseudoviral populations of the most PGT151- and PGT145-reactive virions. Thereby, neutralization by the non-depleting NAb increased, whereas neutralization by the depleting NAb decreased. Furthermore, depletion by PGT151 increased sensitivity to autologous neutralization by sera from rabbits immunized with soluble native-like CZA97.012 trimer: substantial persistent fractions were reduced. NAbs in these sera target epitopes comprising residue D411 at the V4-β19 transition in a defect of the glycan shield on CZA97.012 Env. NAb binding to affinity-fractionated soluble native-like CZA97.012 trimer differed commensurately with neutralization in analyses by ELISA and surface plasmon resonance. Glycan differences between PGT151- and PGT145-purified trimer fractions were then demonstrated by mass spectrometry, providing one explanation for the differential antigenicity. These differences were interpreted in relation to a new structure at 3.4-Å resolution of the soluble CZA97.012 trimer determined by cryo-electron microscopy. The trimer adopted a closed conformation, refuting apex opening as the cause of reduced PGT145 binding to the PGT151-purified form. The evidence suggests that differences in binding and neutralization after trimer purification or pseudovirus depletion with PGT145 or PGT151 are caused by variation in glycosylation, and that some glycan variants affect antigenicity through direct effects on antibody contacts, whereas others act allosterically.
- Department of Microbiology and Immunology, Weill Cornell Medicine, Cornell University, New York, New York, United States of America.
Organizational Affiliation: