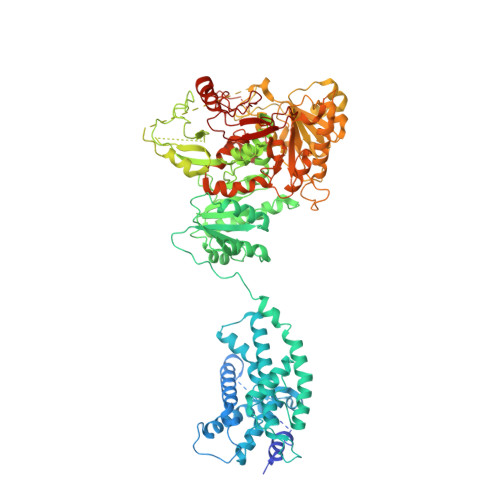
Membrane protein isolation and structure determination in cell-derived membrane vesicles.
Tao, X., Zhao, C., MacKinnon, R.(2023) Proc Natl Acad Sci U S A 120: e2302325120-e2302325120
- PubMed: 37098056
- DOI: https://doi.org/10.1073/pnas.2302325120
- Primary Citation of Related Structures:
8GH9, 8GHF, 8GHG - PubMed Abstract:
Integral membrane protein structure determination traditionally requires extraction from cell membranes using detergents or polymers. Here, we describe the isolation and structure determination of proteins in membrane vesicles derived directly from cells. Structures of the ion channel Slo1 from total cell membranes and from cell plasma membranes were determined at 3.8 Å and 2.7 Å resolution, respectively. The plasma membrane environment stabilizes Slo1, revealing an alteration of global helical packing, polar lipid, and cholesterol interactions that stabilize previously unresolved regions of the channel and an additional ion binding site in the Ca 2+ regulatory domain. The two methods presented enable structural analysis of both internal and plasma membrane proteins without disrupting weakly interacting proteins, lipids, and cofactors that are essential to biological function.
- Laboratory of Molecular Neurobiology and Biophysics, The Rockefeller University, New York, NY 10065.
Organizational Affiliation: