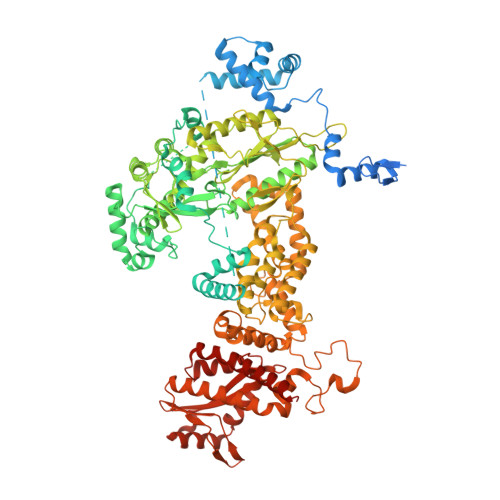
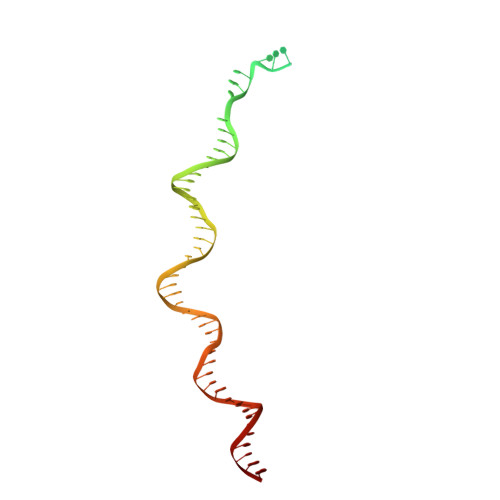
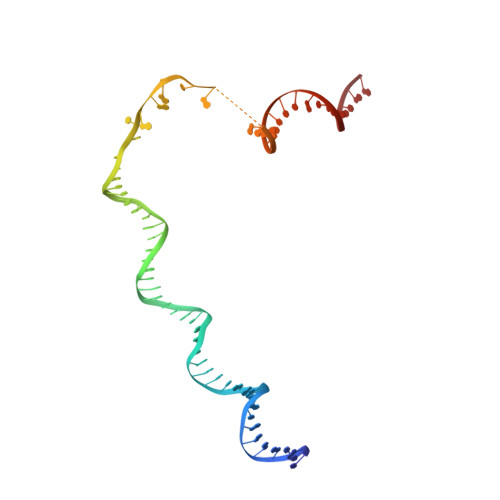
Structure of the R2 non-LTR retrotransposon initiating target-primed reverse transcription.
Wilkinson, M.E., Frangieh, C.J., Macrae, R.K., Zhang, F.(2023) Science 380: 301-308
- PubMed: 37023171
- DOI: https://doi.org/10.1126/science.adg7883
- Primary Citation of Related Structures:
8GH6 - PubMed Abstract:
Non-long terminal repeat (non-LTR) retrotransposons, or long interspersed nuclear elements (LINEs), are an abundant class of eukaryotic transposons that insert into genomes by target-primed reverse transcription (TPRT). During TPRT, a target DNA sequence is nicked and primes reverse transcription of the retrotransposon RNA. Here, we report the cryo-electron microscopy structure of the Bombyx mori R2 non-LTR retrotransposon initiating TPRT at its ribosomal DNA target. The target DNA sequence is unwound at the insertion site and recognized by an upstream motif. An extension of the reverse transcriptase (RT) domain recognizes the retrotransposon RNA and guides the 3' end into the RT active site to template reverse transcription. We used Cas9 to retarget R2 in vitro to non-native sequences, suggesting future use as a reprogrammable RNA-based gene-insertion tool.
- Howard Hughes Medical Institute, Cambridge, MA 02139, USA.
Organizational Affiliation: