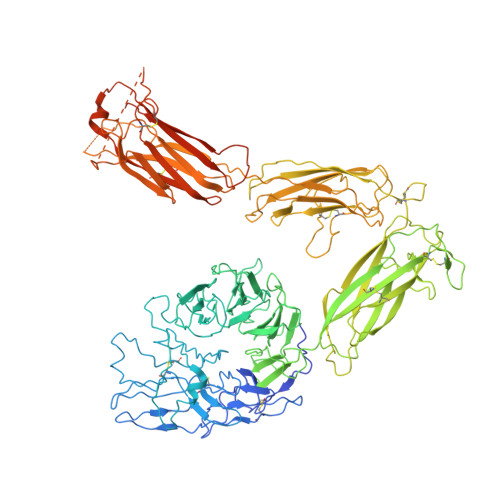
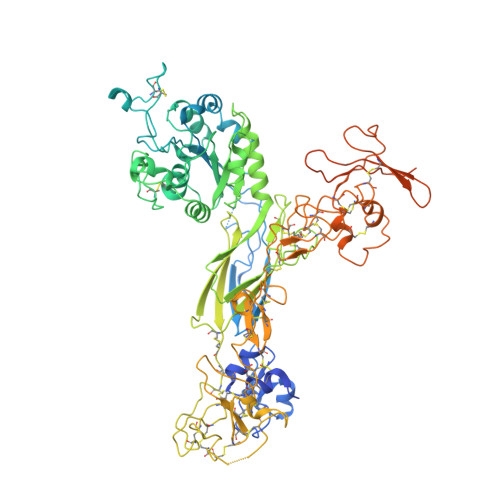
Full-length alpha IIb beta 3 cryo-EM structure reveals intact integrin initiate-activation intrinsic architecture.
Huo, T., Wu, H., Moussa, Z., Sen, M., Dalton, V., Wang, Z.(2024) Structure 32: 899
- PubMed: 38579706
- DOI: https://doi.org/10.1016/j.str.2024.03.006
- Primary Citation of Related Structures:
8GCD, 8GCE - PubMed Abstract:
Integrin αIIbβ3 is the key receptor regulating platelet retraction and accumulation and a proven drug-target for antithrombotic therapies. Here we resolve the cryo-EM structures of the full-length αIIbβ3, which covers three distinct states along the activation pathway. Firstly, we obtain the αIIbβ3 structure at 3 Å resolution in the inactive state, revealing the overall topology of the heterodimer with the transmembrane (TM) helices and the ligand-binding domain tucked in a specific angle proximity to the TM region. After the addition of a Mn 2+ agonist, we resolve two coexisting structures representing two new states between inactive and active state. Our structures show conformational changes of the αIIbβ3 activating trajectory and a unique twisting of the integrin legs, which is required for platelets accumulation. Our structure provides direct structural evidence for how the lower legs are involved in full-length integrin activation mechanisms and offers a new strategy to target the αIIbβ3 lower leg.
- Verna and Marrs McLean Department of Biochemistry and Molecular Biology, Baylor College of Medicine, Houston, TX 77030, USA.
Organizational Affiliation: