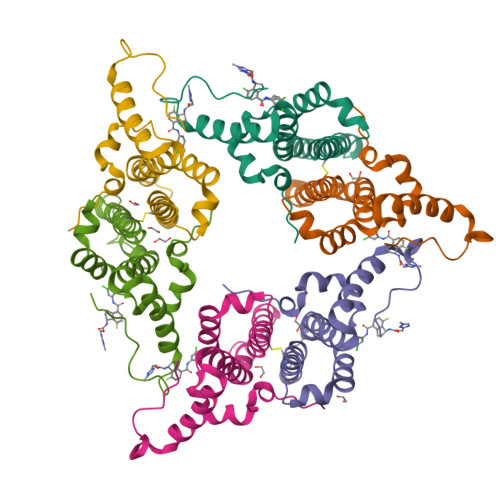
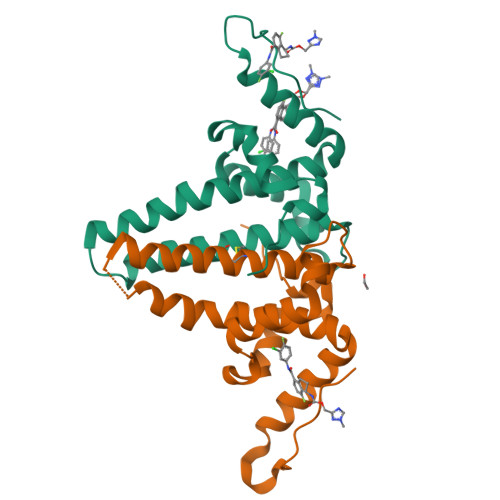
Design, synthesis, and structure-activity relationship of a bicyclic HBV capsid assembly modulator chemotype leading to the identification of clinical candidate AB-506.
Cole, A.G., Kultgen, S.G., Mani, N., Quintero, J.G., Yi Fan, K., Ardzinski, A., Stever, K., Dorsey, B.D., Phelps, J.R., Lee, A.C.H., Thi, E.P., Chiu, T., Tang, S., Horanyi, P.S., Mayclin, S.J., Harasym, T.O., Sofia, M.J.(2023) Bioorg Med Chem Lett 94: 129456-129456
- PubMed: 37633618
- DOI: https://doi.org/10.1016/j.bmcl.2023.129456
- Primary Citation of Related Structures:
8GBU - PubMed Abstract:
Disruption of the HBV capsid assembly process through small-molecule interaction with HBV core protein is a validated target for the suppression of hepatitis B viral replication and the development of new antivirals. Through combination of key structural features associated with two distinct series of capsid assembly modulators, a novel aminochroman-based chemotype was identified. Optimization of anti-HBV potency through generation of SAR in addition to further core modifications provided a series of related functionalized aminoindanes. Key compounds demonstrated excellent cellular potency in addition to favorable ADME and pharmacokinetic profiles and were shown to be highly efficacious in a mouse model of HBV replication. Aminoindane derivative AB-506 was subsequently advanced into clinical development.
Organizational Affiliation:
Arbutus Biopharma, Inc., 701 Veterans Circle, Warminster, PA 18974, USA. Electronic address: acole@arbutusbio.com.