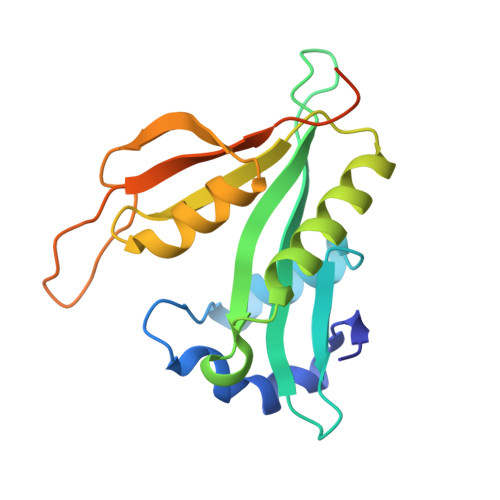
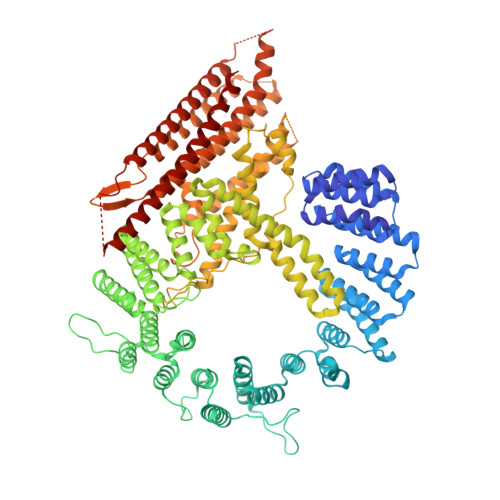
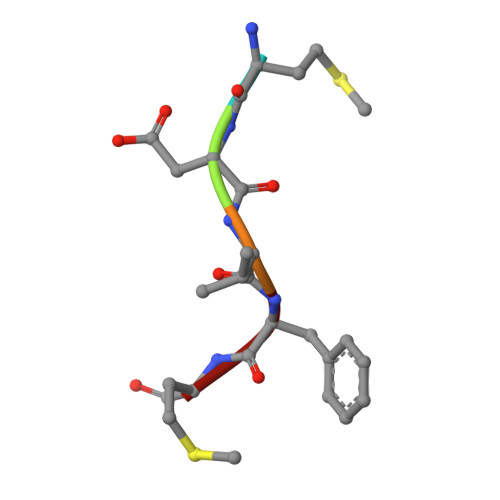
Semi-Synthetic CoA-alpha-Synuclein Constructs Trap N-Terminal Acetyltransferase NatB for Binding Mechanism Studies.
Pan, B., Gardner, S.M., Schultz, K., Perez, R.M., Deng, S., Shimogawa, M., Sato, K., Rhoades, E., Marmorstein, R., Petersson, E.J.(2023) J Am Chem Soc 145: 14019-14030
- PubMed: 37319422
- DOI: https://doi.org/10.1021/jacs.3c03887
- Primary Citation of Related Structures:
8G0L - PubMed Abstract:
N-terminal acetylation is a chemical modification carried out by N-terminal acetyltransferases. A major member of this enzyme family, NatB, acts on much of the human proteome, including α-synuclein (αS), a synaptic protein that mediates vesicle trafficking. NatB acetylation of αS modulates its lipid vesicle binding properties and amyloid fibril formation, which underlies its role in the pathogenesis of Parkinson's disease. Although the molecular details of the interaction between human NatB (hNatB) and the N-terminus of αS have been resolved, whether the remainder of the protein plays a role in interacting with the enzyme is unknown. Here, we execute the first synthesis, by native chemical ligation, of a bisubstrate inhibitor of NatB consisting of coenzyme A and full-length human αS, additionally incorporating two fluorescent probes for studies of conformational dynamics. We use cryo-electron microscopy (cryo-EM) to characterize the structural features of the hNatB/inhibitor complex and show that, beyond the first few residues, αS remains disordered when in complex with hNatB. We further probe changes in the αS conformation by single molecule Förster resonance energy transfer (smFRET) to reveal that the C-terminus expands when bound to hNatB. Computational models based on the cryo-EM and smFRET data help to explain the conformational changes as well as their implications for hNatB substrate recognition and specific inhibition of the interaction with αS. Beyond the study of αS and NatB, these experiments illustrate valuable strategies for the study of challenging structural biology targets through a combination of protein semi-synthesis, cryo-EM, smFRET, and computational modeling.
- Department of Chemistry, University of Pennsylvania, 231 South 34th Street, Philadelphia, Pennsylvania 19104, United States.
Organizational Affiliation: