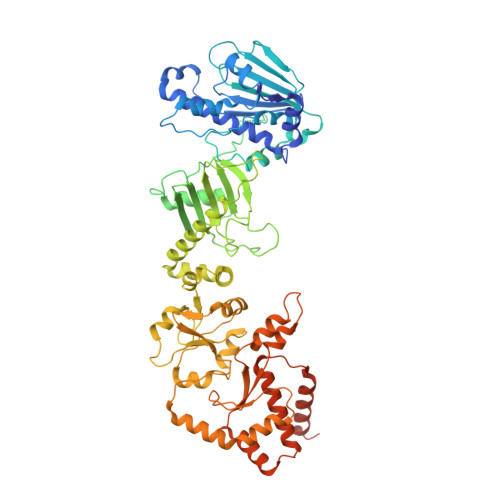
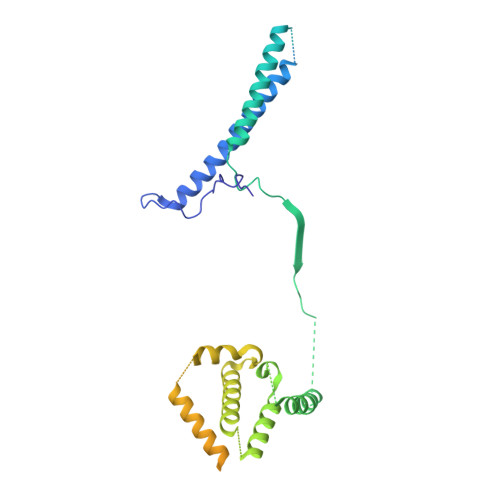
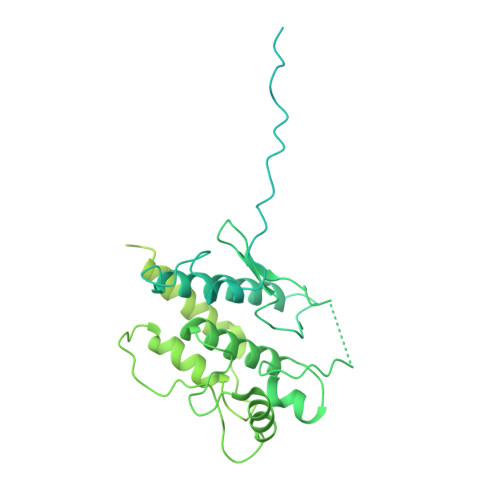
Structural insight into guanylyl cyclase receptor hijacking of the kinase-Hsp90 regulatory mechanism.
Caveney, N.A., Tsutsumi, N., Garcia, K.C.(2023) Elife 12
- PubMed: 37535399
- DOI: https://doi.org/10.7554/eLife.86784
- Primary Citation of Related Structures:
8FX4 - PubMed Abstract:
Membrane receptor guanylyl cyclases play a role in many important facets of human physiology, from regulating blood pressure to intestinal fluid secretion. The structural mechanisms which influence these important physiological processes have yet to be explored. We present the 3.9 Å resolution cryo-EM structure of the human membrane receptor guanylyl cyclase GC-C in complex with Hsp90 and its co-chaperone Cdc37, providing insight into the mechanism of Cdc37 mediated binding of GC-C to the Hsp90 regulatory complex. As a membrane protein and non-kinase client of Hsp90-Cdc37, this work shows the remarkable plasticity of Cdc37 to interact with a broad array of clients with significant sequence variation. Furthermore, this work shows how membrane receptor guanylyl cyclases hijack the regulatory mechanisms used for active kinases to facilitate their regulation. Given the known druggability of Hsp90, these insights can guide the further development of membrane receptor guanylyl cyclase-targeted therapeutics and lead to new avenues to treat hypertension, inflammatory bowel disease, and other membrane receptor guanylyl cyclase-related conditions.
- Departments of Molecular and Cellular Physiology, and Structural Biology, Stanford University School of Medicine, Stanford, United States.
Organizational Affiliation: