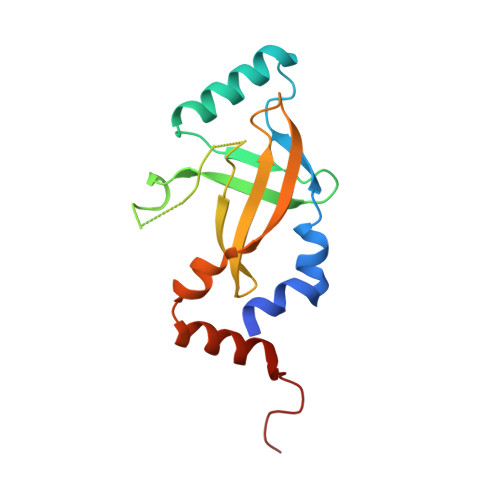
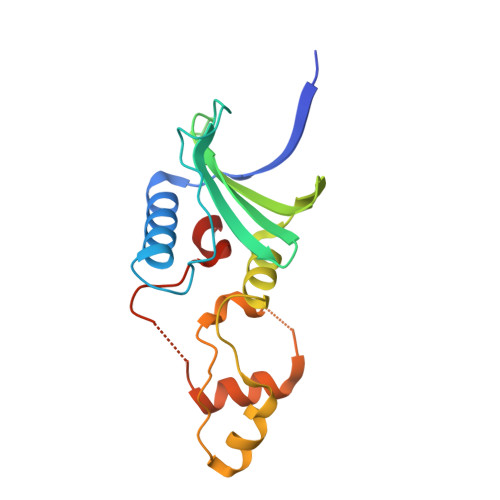
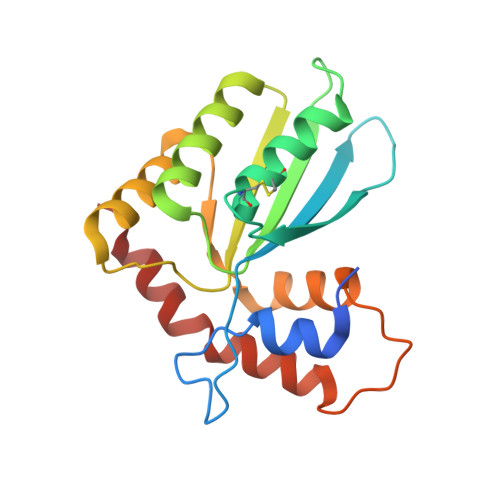
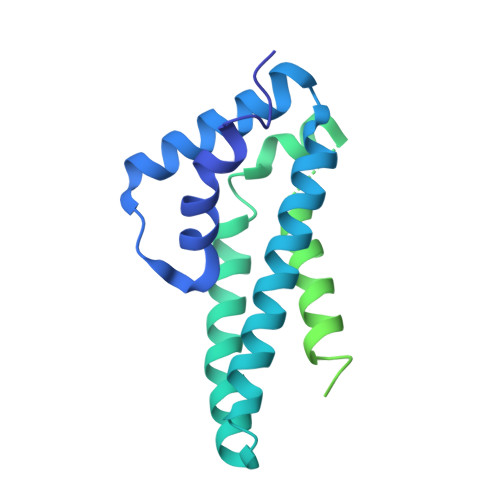
Structural basis of HIV-1 Vif-mediated E3 ligase targeting of host APOBEC3H.
Ito, F., Alvarez-Cabrera, A.L., Kim, K., Zhou, Z.H., Chen, X.S.(2023) Nat Commun 14: 5241-5241
- PubMed: 37640699
- DOI: https://doi.org/10.1038/s41467-023-40955-x
- Primary Citation of Related Structures:
8FVI, 8FVJ - PubMed Abstract:
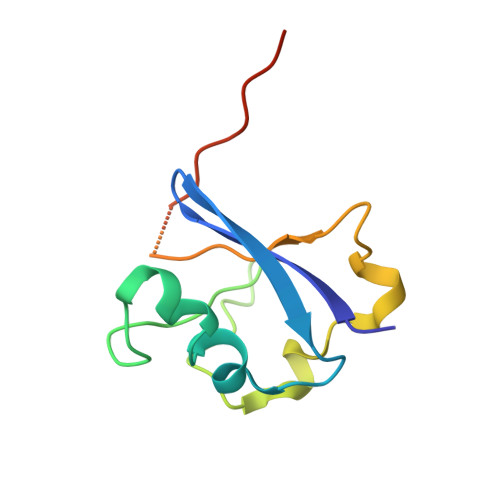
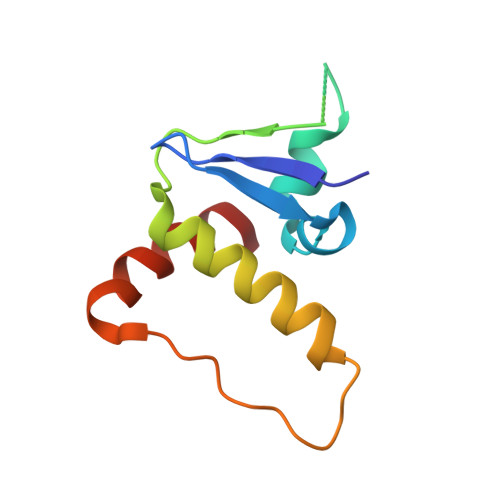
Human APOBEC3 (A3) cytidine deaminases are antiviral factors that are particularly potent against retroviruses. As a countermeasure, HIV-1 uses a viral infectivity factor (Vif) to target specific human A3s for proteasomal degradation. Vif recruits cellular transcription cofactor CBF-β and Cullin-5 (CUL5) RING E3 ubiquitin ligase to bind different A3s distinctively, but how this is accomplished remains unclear in the absence of the atomic structure of the complex. Here, we present the cryo-EM structures of HIV-1 Vif in complex with human A3H, CBF-β and components of CUL5 ubiquitin ligase (CUL5, ELOB, and ELOC). Vif nucleates the entire complex by directly binding four human proteins, A3H, CBF-β, CUL5, and ELOC. The structures reveal a large interface area between A3H and Vif, primarily mediated by an α-helical side of A3H and a five-stranded β-sheet of Vif. This A3H-Vif interface unveils the basis for sensitivity-modulating polymorphism of both proteins, including a previously reported gain-of-function mutation in Vif isolated from HIV/AIDS patients. Our structural and functional results provide insights into the remarkable interplay between HIV and humans and would inform development efforts for anti-HIV therapeutics.
- Molecular and Computational Biology, Department of Biological Sciences, University of Southern California, Los Angeles, CA, 90089, USA.
Organizational Affiliation: