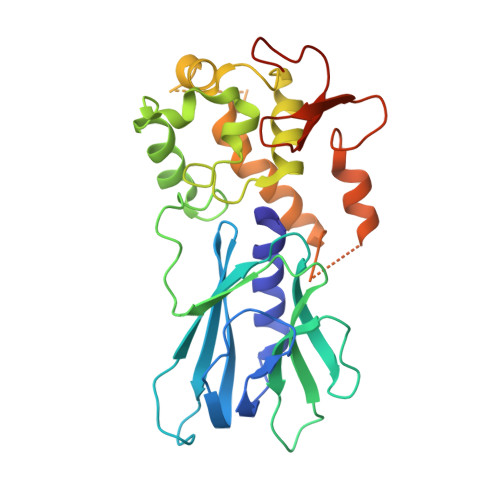
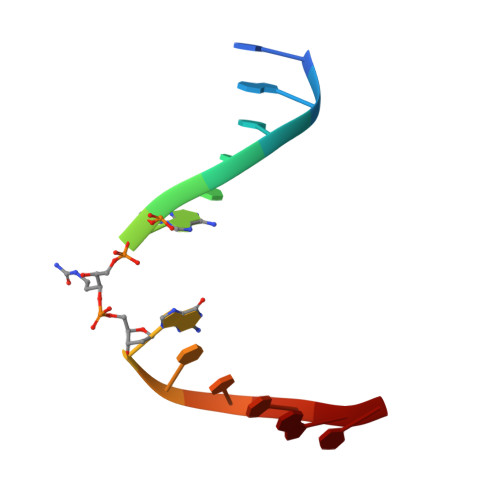
Base excision repair of the N-(2-deoxy-d-erythro-pentofuranosyl)-urea lesion by the hNEIL1 glycosylase.
Tomar, R., Minko, I.G., Sharma, P., Kellum, A.H., Lei, L., Harp, J.M., Iverson, T.M., Lloyd, R.S., Egli, M., Stone, M.P.(2023) Nucleic Acids Res 51: 3754-3769
- PubMed: 37014002
- DOI: https://doi.org/10.1093/nar/gkad164
- Primary Citation of Related Structures:
8FTJ - PubMed Abstract:
The N-(2-deoxy-d-erythro-pentofuranosyl)-urea DNA lesion forms following hydrolytic fragmentation of cis-5R,6S- and trans-5R,6R-dihydroxy-5,6-dihydrothymidine (thymine glycol, Tg) or from oxidation of 7,8-dihydro-8-oxo-deoxyguanosine (8-oxodG) and subsequent hydrolysis. It interconverts between α and β deoxyribose anomers. Synthetic oligodeoxynucleotides containing this adduct are efficiently incised by unedited (K242) and edited (R242) forms of the hNEIL1 glycosylase. The structure of a complex between the active site unedited mutant CΔ100 P2G hNEIL1 (K242) glycosylase and double-stranded (ds) DNA containing a urea lesion reveals a pre-cleavage intermediate, in which the Gly2 N-terminal amine forms a conjugate with the deoxyribose C1' of the lesion, with the urea moiety remaining intact. This structure supports a proposed catalytic mechanism in which Glu3-mediated protonation of O4' facilitates attack at deoxyribose C1'. The deoxyribose is in the ring-opened configuration with the O4' oxygen protonated. The electron density of Lys242 suggests the 'residue 242-in conformation' associated with catalysis. This complex likely arises because the proton transfer steps involving Glu6 and Lys242 are hindered due to Glu6-mediated H-bonding with the Gly2 and the urea lesion. Consistent with crystallographic data, biochemical analyses show that the CΔ100 P2G hNEIL1 (K242) glycosylase exhibits a residual activity against urea-containing dsDNA.
- Department of Chemistry and the Vanderbilt-Ingram Cancer Center, Vanderbilt University, Station B Box 351822, Nashville, TN 37235, USA.
Organizational Affiliation: