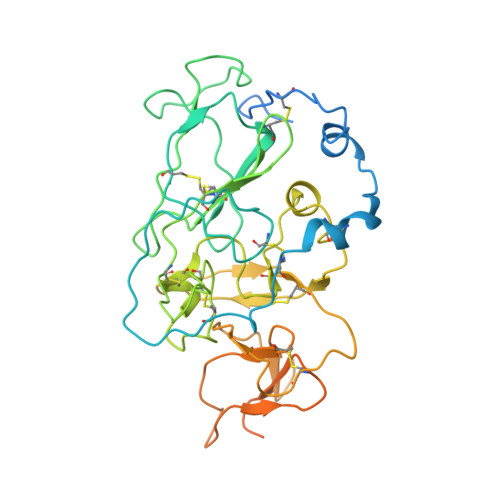
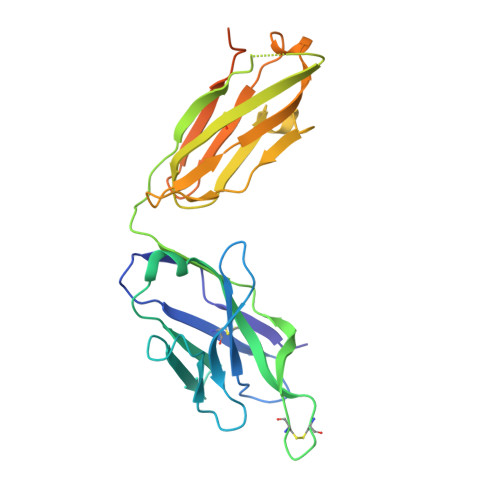
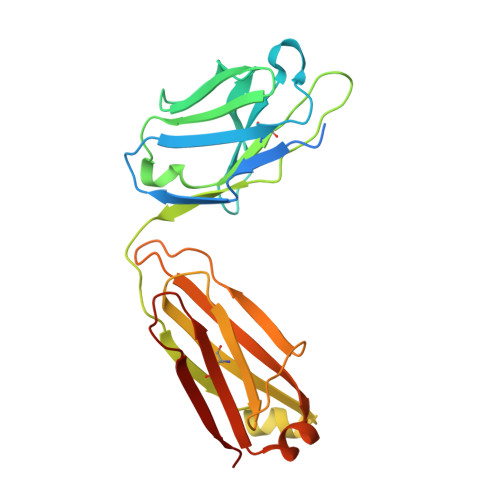
Structure of engineered hepatitis C virus E1E2 ectodomain in complex with neutralizing antibodies.
Metcalf, M.C., Janus, B.M., Yin, R., Wang, R., Guest, J.D., Pozharski, E., Law, M., Mariuzza, R.A., Toth, E.A., Pierce, B.G., Fuerst, T.R., Ofek, G.(2023) Nat Commun 14: 3980-3980
- PubMed: 37407593
- DOI: https://doi.org/10.1038/s41467-023-39659-z
- Primary Citation of Related Structures:
8FSJ - PubMed Abstract:
Hepatitis C virus (HCV) is a major global health burden as the leading causative agent of chronic liver disease and hepatocellular carcinoma. While the main antigenic target for HCV-neutralizing antibodies is the membrane-associated E1E2 surface glycoprotein, the development of effective vaccines has been hindered by complications in the biochemical preparation of soluble E1E2 ectodomains. Here, we present a cryo-EM structure of an engineered, secreted E1E2 ectodomain of genotype 1b in complex with neutralizing antibodies AR4A, HEPC74, and IGH520. Structural characterization of the E1 subunit and C-terminal regions of E2 reveal an overall architecture of E1E2 that concurs with that observed for non-engineered full-length E1E2. Analysis of the AR4A epitope within a region of E2 that bridges between the E2 core and E1 defines the structural basis for its broad neutralization. Our study presents the structure of an E1E2 complex liberated from membrane via a designed scaffold, one that maintains all essential structural features of native E1E2. The study advances the understanding of the E1E2 heterodimer structure, crucial for the rational design of secreted E1E2 antigens in vaccine development.
- Department of Cell Biology and Molecular Genetics, University of Maryland, College Park, MD, USA.
Organizational Affiliation: