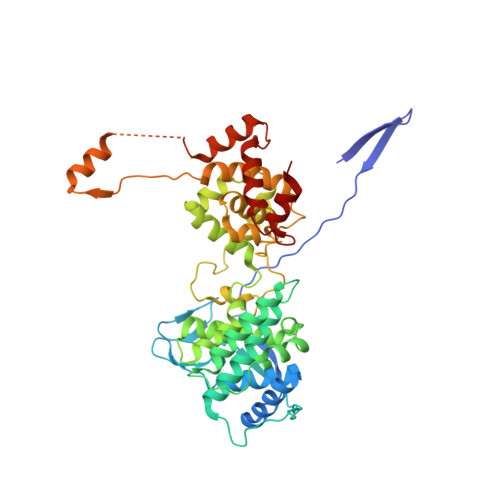
Structure and Dynamics of the Unassembled Nucleoprotein of Rabies Virus in Complex with Its Phosphoprotein Chaperone Module.
Gerard, F.C.A., Bourhis, J.M., Mas, C., Branchard, A., Vu, D.D., Varhoshkova, S., Leyrat, C., Jamin, M.(2022) Viruses 14
- PubMed: 36560817
- DOI: https://doi.org/10.3390/v14122813
- Primary Citation of Related Structures:
8B8V, 8FFR - PubMed Abstract:
As for all non-segmented negative RNA viruses, rabies virus has its genome packaged in a linear assembly of nucleoprotein (N), named nucleocapsid. The formation of new nucleocapsids during virus replication in cells requires the production of soluble N protein in complex with its phosphoprotein (P) chaperone. In this study, we reconstituted a soluble heterodimeric complex between an armless N protein of rabies virus (RABV), lacking its N-terminal subdomain (N NT-ARM ), and a peptide encompassing the N 0 chaperon module of the P protein. We showed that the chaperone module undergoes a disordered-order transition when it assembles with N 0 and measured an affinity in the low nanomolar range using a competition assay. We solved the crystal structure of the complex at a resolution of 2.3 Å, unveiling the details of the conserved interfaces. MD simulations showed that both the chaperon module of P and RNA-mediated polymerization reduced the ability of the RNA binding cavity to open and close. Finally, by reconstituting a complex with full-length P protein, we demonstrated that each P dimer could independently chaperon two N 0 molecules.
- Institut de Biologie Structurale (IBS), Université Grenoble Alpes, CEA, CNRS, 71 Avenue des Martyrs, 38000 Grenoble, France.
Organizational Affiliation: