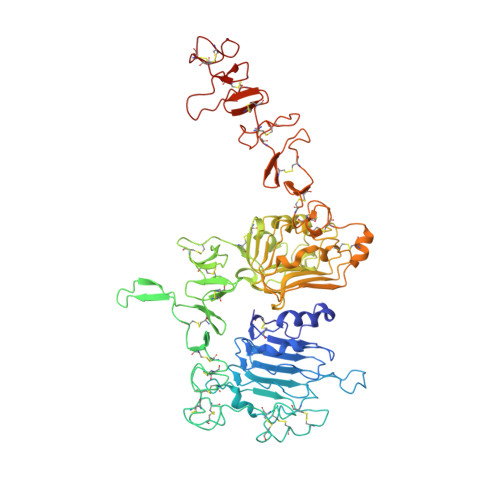
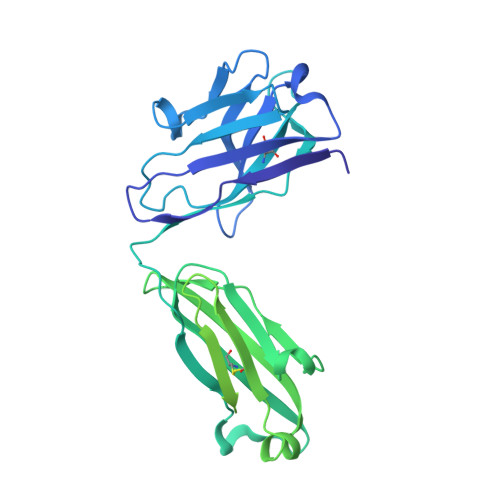
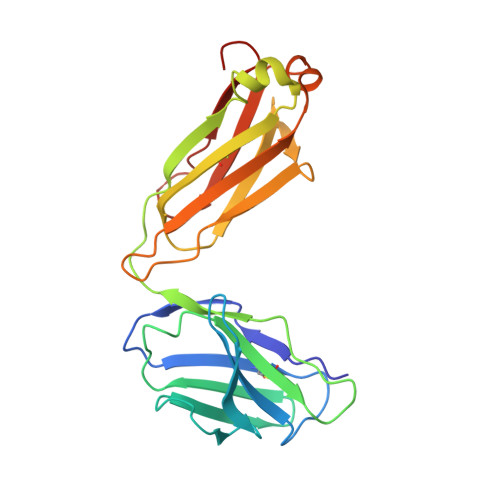
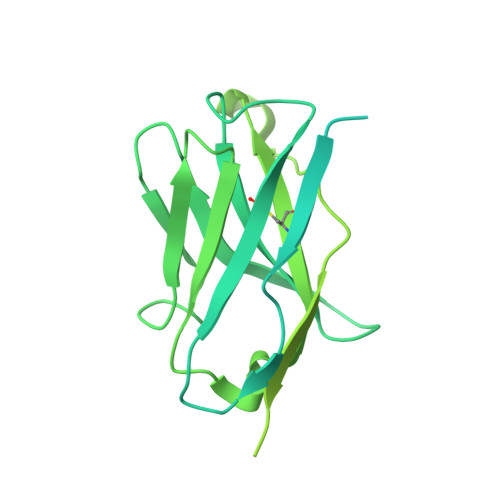
An anti-HER2 biparatopic antibody that induces unique HER2 clustering and complement-dependent cytotoxicity.
Weisser, N.E., Sanches, M., Escobar-Cabrera, E., O'Toole, J., Whalen, E., Chan, P.W.Y., Wickman, G., Abraham, L., Choi, K., Harbourne, B., Samiotakis, A., Rojas, A.H., Volkers, G., Wong, J., Atkinson, C.E., Baardsnes, J., Worrall, L.J., Browman, D., Smith, E.E., Baichoo, P., Cheng, C.W., Guedia, J., Kang, S., Mukhopadhyay, A., Newhook, L., Ohrn, A., Raghunatha, P., Zago-Schmitt, M., Schrag, J.D., Smith, J., Zwierzchowski, P., Scurll, J.M., Fung, V., Black, S., Strynadka, N.C.J., Gold, M.R., Presta, L.G., Ng, G., Dixit, S.(2023) Nat Commun 14: 1394-1394
- PubMed: 36914633
- DOI: https://doi.org/10.1038/s41467-023-37029-3
- Primary Citation of Related Structures:
8FFJ - PubMed Abstract:
Human epidermal growth factor receptor 2 (HER2) is a receptor tyrosine kinase that plays an oncogenic role in breast, gastric and other solid tumors. However, anti-HER2 therapies are only currently approved for the treatment of breast and gastric/gastric esophageal junction cancers and treatment resistance remains a problem. Here, we engineer an anti-HER2 IgG1 bispecific, biparatopic antibody (Ab), zanidatamab, with unique and enhanced functionalities compared to both trastuzumab and the combination of trastuzumab plus pertuzumab (tras + pert). Zanidatamab binds adjacent HER2 molecules in trans and initiates distinct HER2 reorganization, as shown by polarized cell surface HER2 caps and large HER2 clusters, not observed with trastuzumab or tras + pert. Moreover, zanidatamab, but not trastuzumab nor tras + pert, elicit potent complement-dependent cytotoxicity (CDC) against high HER2-expressing tumor cells in vitro. Zanidatamab also mediates HER2 internalization and downregulation, inhibition of both cell signaling and tumor growth, antibody-dependent cellular cytotoxicity (ADCC) and phagocytosis (ADCP), and also shows superior in vivo antitumor activity compared to tras + pert in a HER2-expressing xenograft model. Collectively, we show that zanidatamab has multiple and distinct mechanisms of action derived from the structural effects of biparatopic HER2 engagement.
- Zymeworks BC Inc., 114 East 4th Avenue, Suite 800, Vancouver, BC, Canada. nweisser@zymeworks.com.
Organizational Affiliation: