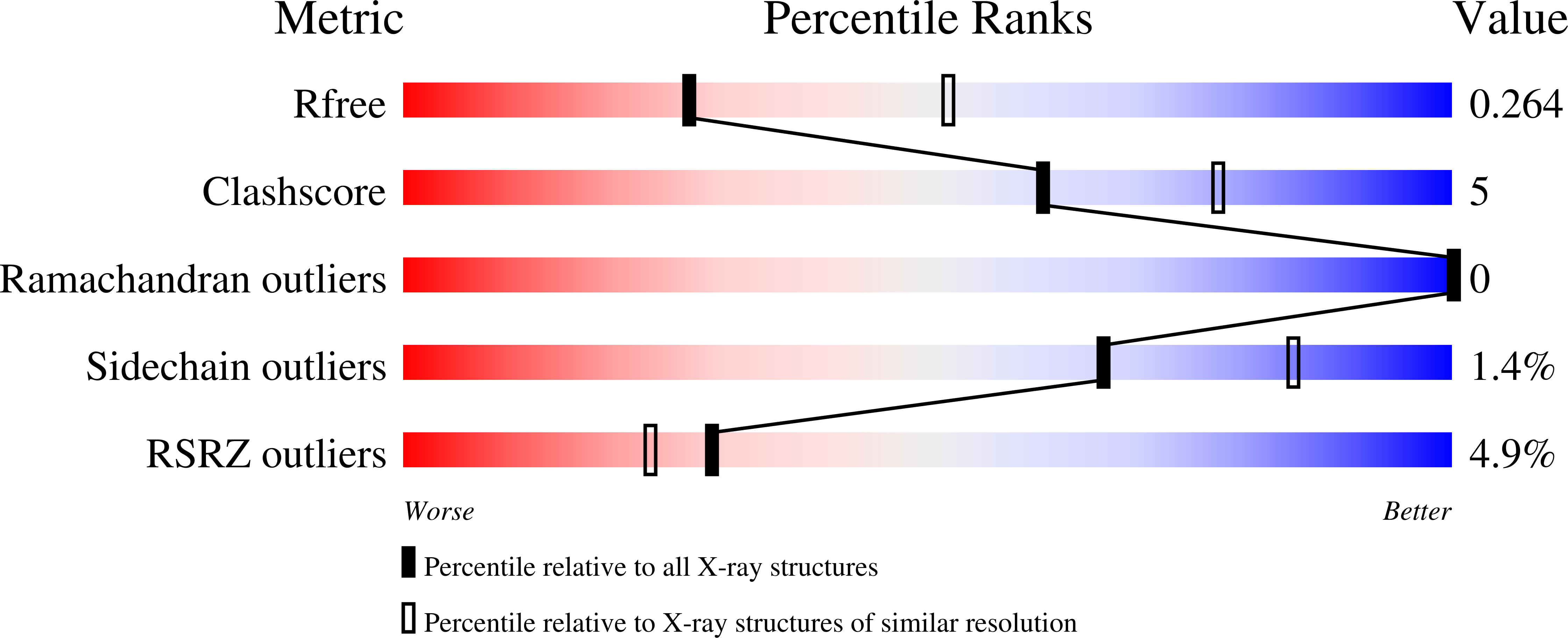
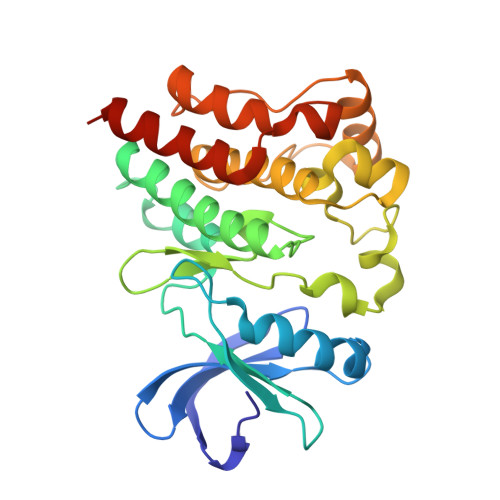
Structure of BTK kinase domain with the second-generation inhibitors acalabrutinib and tirabrutinib.
Lin, D.Y., Andreotti, A.H.(2023) PLoS One 18: e0290872-e0290872
- PubMed: 37651403
- DOI: https://doi.org/10.1371/journal.pone.0290872
- Primary Citation of Related Structures:
8FD9, 8FF0 - PubMed Abstract:
Bruton's tyrosine kinase (BTK) is the target of the therapeutic agent, Ibrutinib, that treats chronic lymphocyte leukemia (CLL), mantle cell lymphoma (MCL) and other B cell malignancies. Ibrutinib is a first in class, covalent BTK inhibitor that limits B-cell survival and proliferation. Designing new inhibitors of BTK has been an important objective for advancing development of improved therapeutic agents against cancer and autoimmune disorders. Based on the success of Ibrutinib, several second-generation irreversible BTK inhibitors have been developed that exhibit fewer off-target effects. However, the binding-mode and their interaction with Btk have not been experimentally determined and evaluated at atomic resolution. Here we determined the first crystal structure of the BTK kinase domain in complex with acalabrutinib. In addition, we report a structure of the BTK/tirabrutinib complex and compare these structures with previously solved structures. The structures provide insight in the superior selectivity reported for acalabrutinb and guide future BTK inhibitor development.
Organizational Affiliation:
Roy J. Carver Department of Biochemistry, Biophysics and Molecular Biology, Iowa State, University, Ames, IA, United States of America.