Biochemical Studies of Systemic Lupus Erythematosus-Associated Mutations in Nonreceptor Tyrosine Kinases Ack1 and Brk.
Kan, Y., Paung, Y., Kim, Y., Seeliger, M.A., Miller, W.T.(2023) Biochemistry 62: 1124-1137
- PubMed: 36854171
- DOI: https://doi.org/10.1021/acs.biochem.2c00685
- Primary Citation of Related Structures:
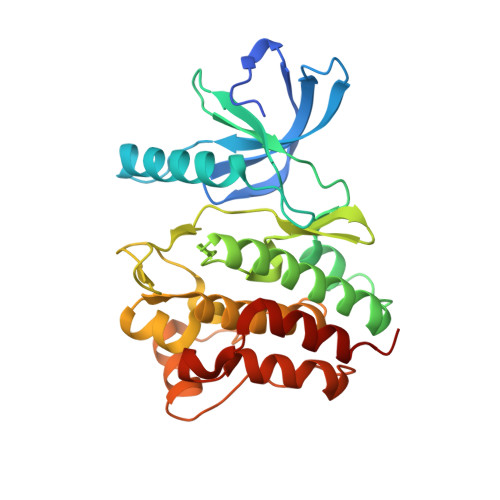
8FE9 - PubMed Abstract:
Tyrosine kinases (TKs) play essential roles in signaling processes that regulate cell survival, migration, and proliferation. Dysregulation of tyrosine kinases underlies many disorders, including cancer, cardiovascular and developmental diseases, as well as pathologies of the immune system. Ack1 and Brk are nonreceptor tyrosine kinases (NRTKs) best known for their roles in cancer. Here, we have biochemically characterized novel Ack1 and Brk mutations identified in patients with systemic lupus erythematosus (SLE). These mutations are the first SLE-linked polymorphisms found among NRTKs. We show that two of the mutants are catalytically inactive, while the other three have reduced activity. To understand the structural changes associated with the loss-of-function phenotype, we solved the crystal structure of one of the Ack1 kinase mutants, K161Q. Furthermore, two of the mutated residues (Ack1 A156 and K161) critical for catalytic activity are highly conserved among other TKs, and their substitution in other members of the kinase family could have implications in cancer. In contrast to canonical gain-of-function mutations in TKs observed in many cancers, we report loss-of-function mutations in Ack1 and Brk, highlighting the complexity of TK involvement in human diseases.
- Department of Physiology and Biophysics, School of Medicine, Stony Brook University, Stony Brook, New York 11794-8661, United States.
Organizational Affiliation: