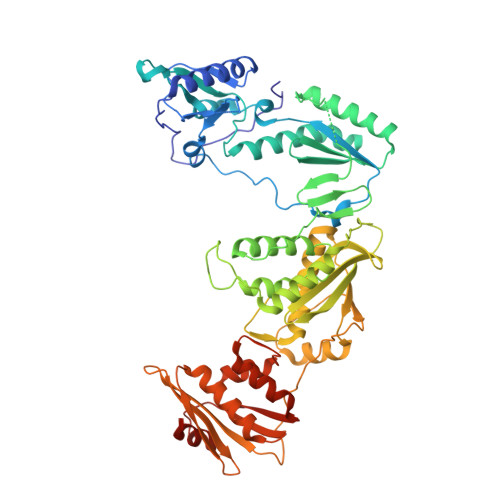
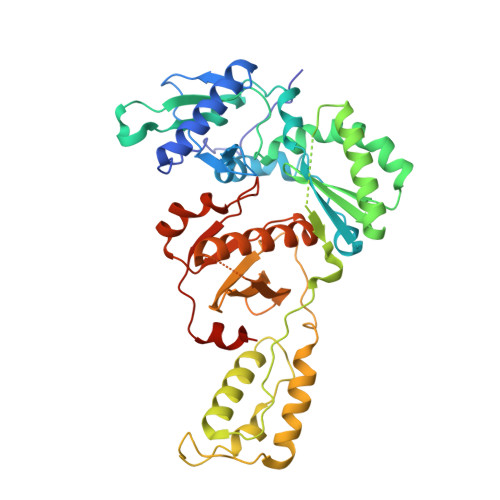
Design and Synthesis of Novel HIV-1 NNRTIs with Bicyclic Cores and with Improved Physicochemical Properties.
Prener, L., Baszczynski, O., Kaiser, M.M., Dracinsky, M., Stepan, G., Lee, Y.J., Brumshtein, B., Yu, H., Jansa, P., Lansdon, E.B., Janeba, Z.(2023) J Med Chem 66: 1761-1777
- PubMed: 36652602
- DOI: https://doi.org/10.1021/acs.jmedchem.2c01574
- Primary Citation of Related Structures:
8FCC, 8FCD, 8FCE - PubMed Abstract:
Non-nucleoside reverse transcriptase inhibitors (NNRTIs) represent cornerstones of current regimens for treatment of human immunodeficiency virus type 1 (HIV-1) infections. However, NNRTIs usually suffer from low aqueous solubility and the emergence of resistant viral strains. In the present work, novel bicyclic NNRTIs derived from etravirine (ETV) and rilpivirine (RPV), bearing modified purine, tetrahydropteridine, and pyrimidodiazepine cores, were designed and prepared. Compounds 2 , 4 , and 6 carrying the acrylonitrile moiety displayed single-digit nanomolar activities against the wild-type (WT) virus (EC 50 = 2.5, 2.7, and 3.0 nM, respectively), where the low nanomolar activity was retained against HXB2 (EC 50 = 2.2-2.8 nM) and the K103N and Y181C mutated strains (fold change, 1.2-6.7×). Most importantly, compound 2 exhibited significantly improved phosphate-buffered saline solubility (10.4 μM) compared to ETV and RPV (≪1 μM). Additionally, the binding modes of compounds 2 , 4 , and 6 to the reverse transcriptase were studied by X-ray crystallography.
- Institute of Organic Chemistry and Biochemistry of the Czech Academy of Sciences, Flemingovo nám. 2, Prague 6 160 00, Czech Republic.
Organizational Affiliation: