The structure of Arabidopsis phytochrome A reveals topological and functional diversification among the plant photoreceptor isoforms.
Burgie, E.S., Li, H., Gannam, Z.T.K., McLoughlin, K.E., Vierstra, R.D., Li, H.(2023) Nat Plants 9: 1116-1129
- PubMed: 37291396
- DOI: https://doi.org/10.1038/s41477-023-01435-8
- Primary Citation of Related Structures:
8F5Z - PubMed Abstract:
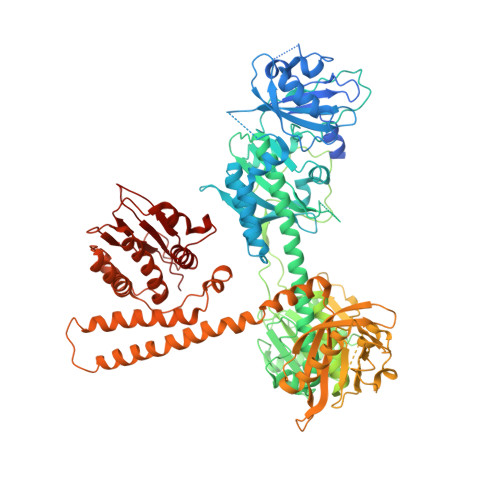
Plants employ a divergent cohort of phytochrome (Phy) photoreceptors to govern many aspects of morphogenesis through reversible photointerconversion between inactive Pr and active Pfr conformers. The two most influential are PhyA whose retention of Pfr enables sensation of dim light, while the relative instability of Pfr for PhyB makes it better suited for detecting full sun and temperature. To better understand these contrasts, we solved, by cryo-electron microscopy, the three-dimensional structure of full-length PhyA as Pr. Like PhyB, PhyA dimerizes through head-to-head assembly of its C-terminal histidine kinase-related domains (HKRDs), while the remainder assembles as a head-to-tail light-responsive platform. Whereas the platform and HKRDs associate asymmetrically in PhyB dimers, these lopsided connections are absent in PhyA. Analysis of truncation and site-directed mutants revealed that this decoupling and altered platform assembly have functional consequences for Pfr stability of PhyA and highlights how plant Phy structural diversification has extended light and temperature perception.
- Department of Biology, Washington University in St. Louis, St. Louis, MO, USA.
Organizational Affiliation: