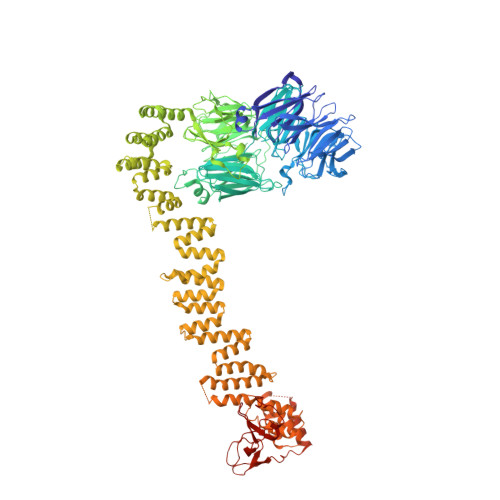
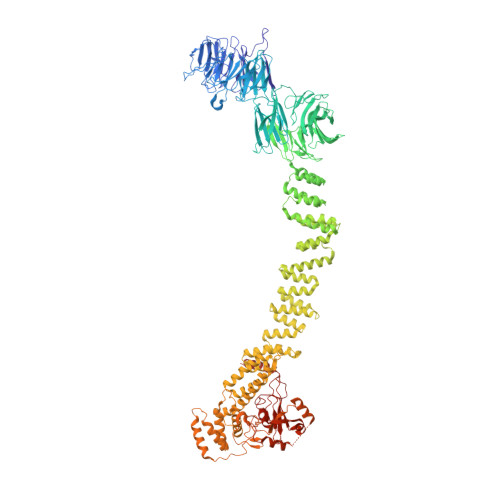
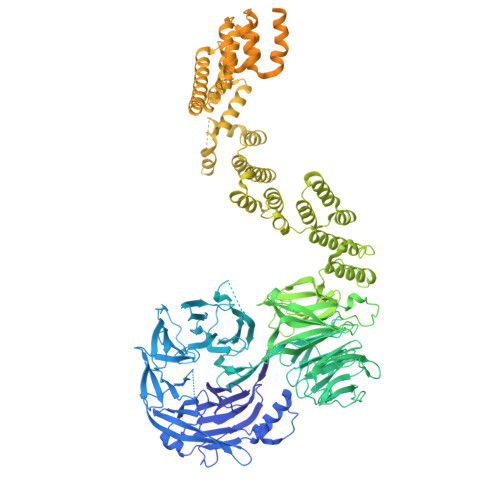
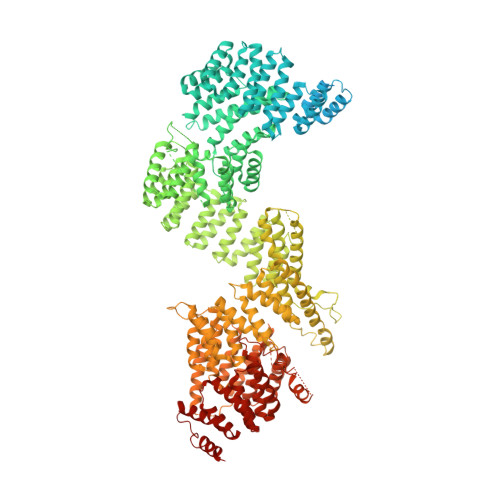
Mechanism of IFT-A polymerization into trains for ciliary transport.
Meleppattu, S., Zhou, H., Dai, J., Gui, M., Brown, A.(2022) Cell 185: 4986
- PubMed: 36563665
- DOI: https://doi.org/10.1016/j.cell.2022.11.033
- Primary Citation of Related Structures:
8F5O, 8F5P - PubMed Abstract:
Intraflagellar transport (IFT) is the highly conserved process by which proteins are transported along ciliary microtubules by a train-like polymeric assembly of IFT-A and IFT-B complexes. IFT-A is sandwiched between IFT-B and the ciliary membrane, consistent with its putative role in transporting transmembrane and membrane-associated cargoes. Here, we have used single-particle analysis electron cryomicroscopy (cryo-EM) to determine structures of native IFT-A complexes. We show that subcomplex rearrangements enable IFT-A to polymerize laterally on anterograde IFT trains, revealing a cooperative assembly mechanism. Surprisingly, we discover that binding of IFT-A to IFT-B shields the preferred lipid-binding interface from the ciliary membrane but orients an interconnected network of β-propeller domains with the capacity to accommodate diverse cargoes toward the ciliary membrane. This work provides a mechanistic basis for understanding IFT-train assembly and cargo interactions.
- Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, 240 Longwood Avenue, Boston, MA 02115, USA.
Organizational Affiliation: