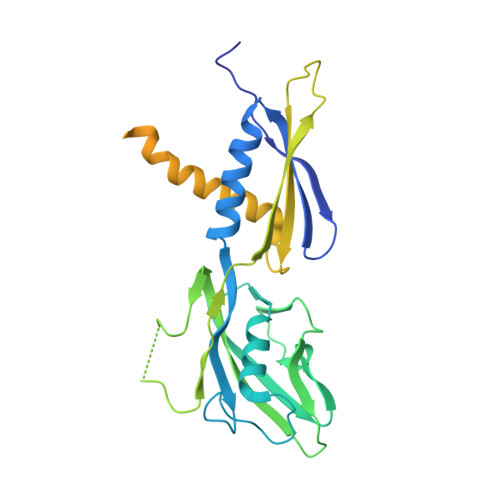
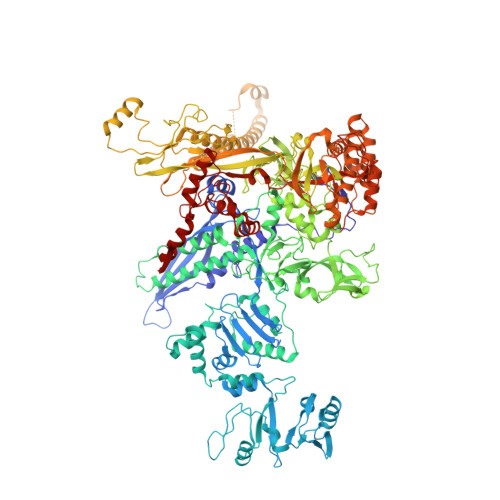
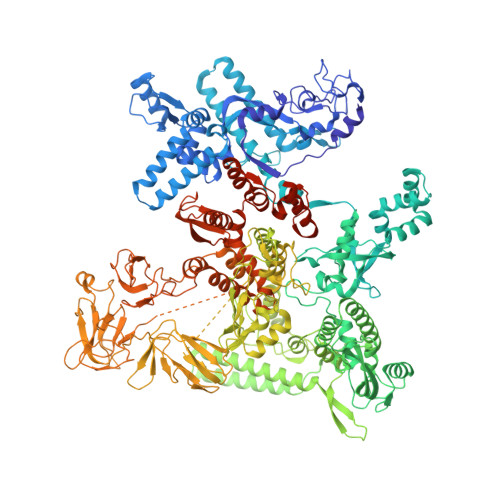
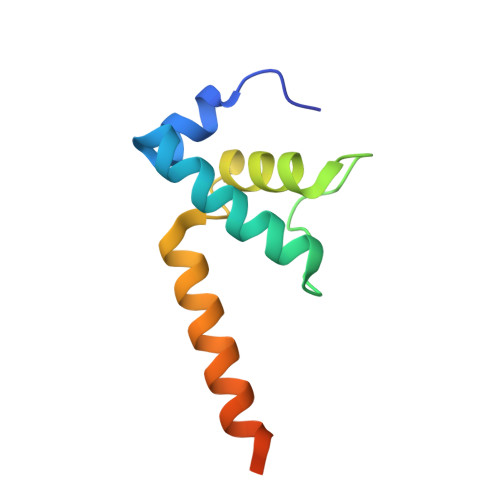
A general mechanism for transcription bubble nucleation in bacteria.
Mueller, A.U., Chen, J., Wu, M., Chiu, C., Nixon, B.T., Campbell, E.A., Darst, S.A.(2023) Proc Natl Acad Sci U S A 120: e2220874120-e2220874120
- PubMed: 36972428
- DOI: https://doi.org/10.1073/pnas.2220874120
- Primary Citation of Related Structures:
8F1I, 8F1J, 8F1K - PubMed Abstract:
Bacterial transcription initiation requires σ factors for nucleation of the transcription bubble. The canonical housekeeping σ factor, σ 70 , nucleates DNA melting via recognition of conserved bases of the promoter -10 motif, which are unstacked and captured in pockets of σ 70 . By contrast, the mechanism of transcription bubble nucleation and formation during the unrelated σ N -mediated transcription initiation is poorly understood. Herein, we combine structural and biochemical approaches to establish that σ N , like σ 70 , captures a flipped, unstacked base in a pocket formed between its N-terminal region I (RI) and extra-long helix features. Strikingly, RI inserts into the nascent bubble to stabilize the nucleated bubble prior to engagement of the obligate ATPase activator. Our data suggest a general paradigm of transcription initiation that requires σ factors to nucleate an early melted intermediate prior to productive RNA synthesis.
- Laboratory of Molecular Biophysics, The Rockefeller University, New York, NY 10065.
Organizational Affiliation: