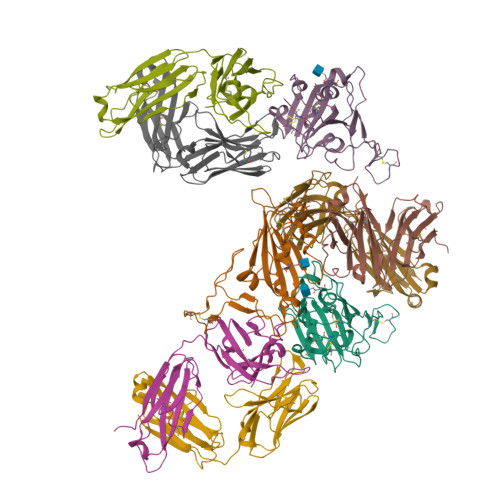
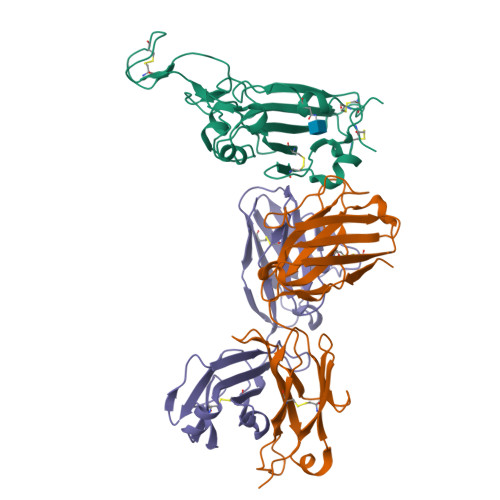
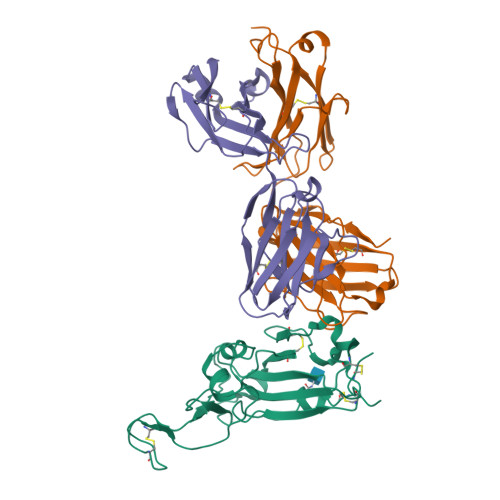
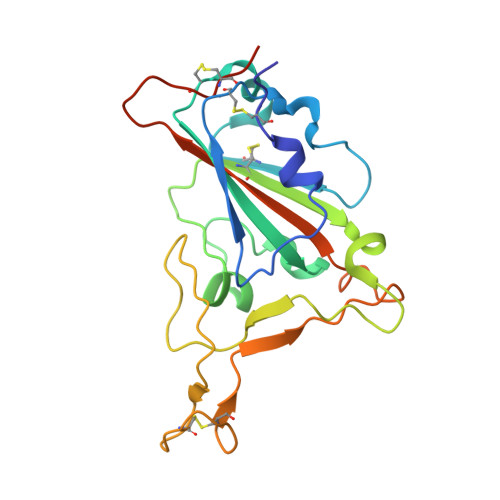
Broad SARS-CoV-2 neutralization by monoclonal and bispecific antibodies derived from a Gamma-infected individual.
Guerra, D., Beaumont, T., Radic, L., Kerster, G., van der Straten, K., Yuan, M., Torres, J.L., Lee, W.H., Liu, H., Poniman, M., Bontjer, I., Burger, J.A., Claireaux, M., Caniels, T.G., Snitselaar, J.L., Bijl, T.P.L., Kruijer, S., Ozorowski, G., Gideonse, D., Sliepen, K., Ward, A.B., Eggink, D., de Bree, G.J., Wilson, I.A., Sanders, R.W., van Gils, M.J.(2023) iScience 26: 108009-108009
- PubMed: 37841584
- DOI: https://doi.org/10.1016/j.isci.2023.108009
- Primary Citation of Related Structures:
8F0I - PubMed Abstract:
The pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has remained a medical threat due to the evolution of multiple variants that acquire resistance to vaccines and prior infection. Therefore, it is imperative to discover monoclonal antibodies (mAbs) that neutralize a broad range of SARS-CoV-2 variants. A stabilized spike glycoprotein was used to enrich antigen-specific B cells from an individual with a primary Gamma variant infection. Five mAbs selected from those B cells showed considerable neutralizing potency against multiple variants, with COVA309-35 being the most potent against the autologous virus, as well as Omicron BA.1 and BA.2, and COVA309-22 having binding and neutralization activity against Omicron BA.4/5, BQ.1.1, and XBB.1. When combining the COVA309 mAbs as cocktails or bispecific antibodies, the breadth and potency were improved. In addition, the mechanism of cross-neutralization of the COVA309 mAbs was elucidated by structural analysis. Altogether these data indicate that a Gamma-infected individual can develop broadly neutralizing antibodies.
Organizational Affiliation:
Amsterdam UMC, location University of Amsterdam, Department of Medical Microbiology and Infection Prevention, Laboratory of Experimental Virology, Meibergdreef 9, Amsterdam 1105 AZ, the Netherlands.