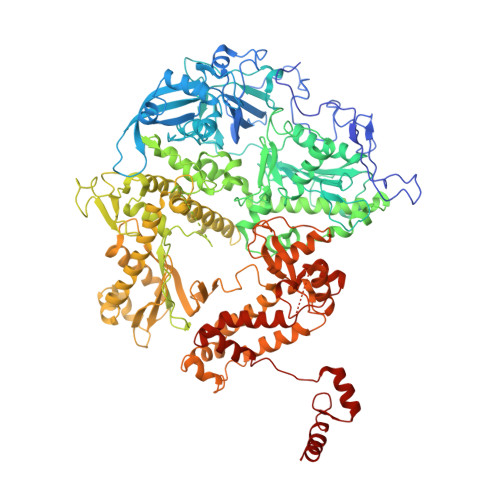
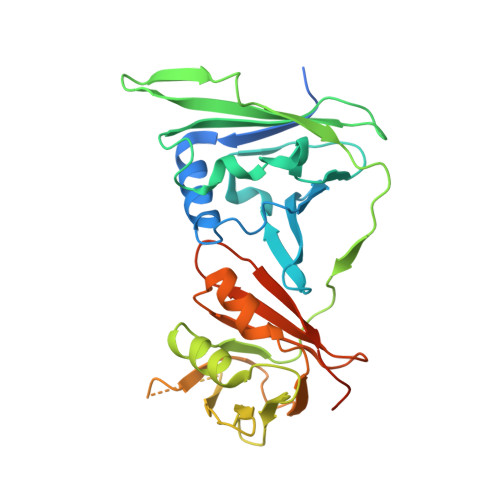
Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Shankar, S., Pan, J., Yang, P., Bian, Y., Oroszlan, G., Yu, Z., Mukherjee, P., Filman, D.J., Hogle, J.M., Shekhar, M., Coen, D.M., Abraham, J.(2024) Cell 187: 5572-5586.e15
- PubMed: 39197451
- DOI: https://doi.org/10.1016/j.cell.2024.07.048
- Primary Citation of Related Structures:
8EXX, 8V1Q, 8V1R, 8V1S, 8V1T - PubMed Abstract:
DNA polymerases are important drug targets, and many structural studies have captured them in distinct conformations. However, a detailed understanding of the impact of polymerase conformational dynamics on drug resistance is lacking. We determined cryoelectron microscopy (cryo-EM) structures of DNA-bound herpes simplex virus polymerase holoenzyme in multiple conformations and interacting with antivirals in clinical use. These structures reveal how the catalytic subunit Pol and the processivity factor UL42 bind DNA to promote processive DNA synthesis. Unexpectedly, in the absence of an incoming nucleotide, we observed Pol in multiple conformations with the closed state sampled by the fingers domain. Drug-bound structures reveal how antivirals may selectively bind enzymes that more readily adopt the closed conformation. Molecular dynamics simulations and the cryo-EM structure of a drug-resistant mutant indicate that some resistance mutations modulate conformational dynamics rather than directly impacting drug binding, thus clarifying mechanisms that drive drug selectivity.
- Department of Microbiology, Blavatnik Institute, Harvard Medical School, Boston, MA 02115, USA.
Organizational Affiliation: