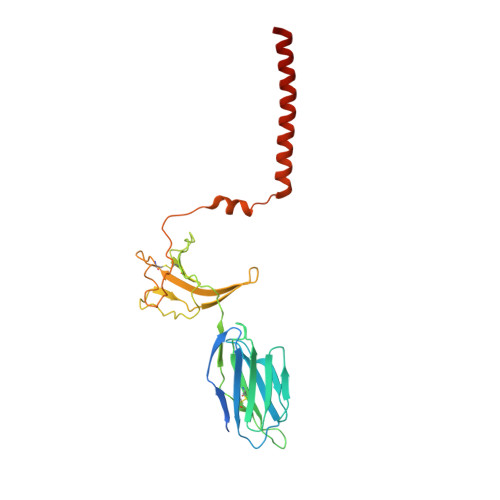
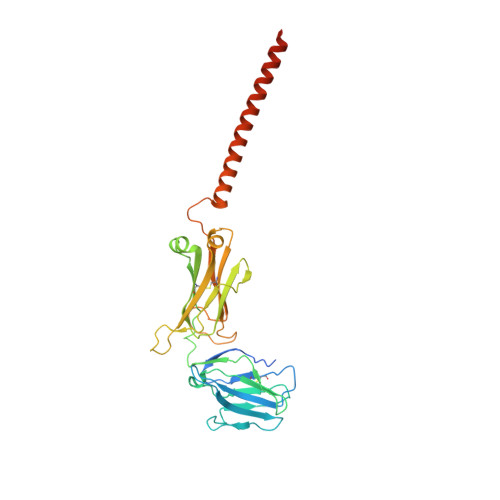
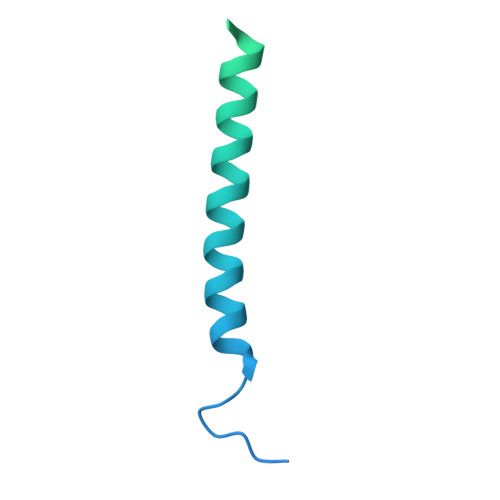
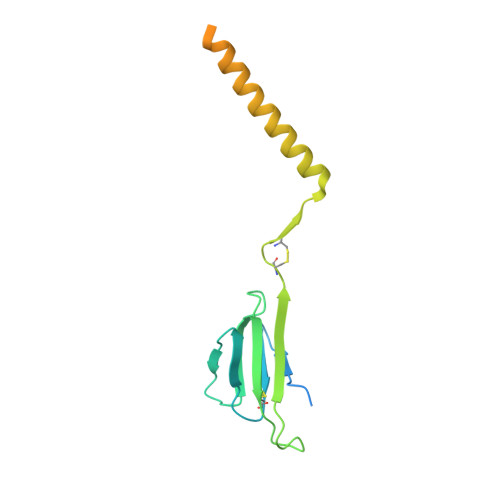
Structural analysis of cancer-relevant TCR-CD3 and peptide-MHC complexes by cryoEM.
Saotome, K., Dudgeon, D., Colotti, K., Moore, M.J., Jones, J., Zhou, Y., Rafique, A., Yancopoulos, G.D., Murphy, A.J., Lin, J.C., Olson, W.C., Franklin, M.C.(2023) Nat Commun 14: 2401-2401
- PubMed: 37100770
- DOI: https://doi.org/10.1038/s41467-023-37532-7
- Primary Citation of Related Structures:
8ES7, 8ES8, 8ES9, 8ESA, 8ESB - PubMed Abstract:
The recognition of antigenic peptide-MHC (pMHC) molecules by T-cell receptors (TCR) initiates the T-cell mediated immune response. Structural characterization is key for understanding the specificity of TCR-pMHC interactions and informing the development of therapeutics. Despite the rapid rise of single particle cryoelectron microscopy (cryoEM), x-ray crystallography has remained the preferred method for structure determination of TCR-pMHC complexes. Here, we report cryoEM structures of two distinct full-length α/β TCR-CD3 complexes bound to their pMHC ligand, the cancer-testis antigen HLA-A2/MAGEA4 (230-239). We also determined cryoEM structures of pMHCs containing MAGEA4 (230-239) peptide and the closely related MAGEA8 (232-241) peptide in the absence of TCR, which provided a structural explanation for the MAGEA4 preference displayed by the TCRs. These findings provide insights into the TCR recognition of a clinically relevant cancer antigen and demonstrate the utility of cryoEM for high-resolution structural analysis of TCR-pMHC interactions.
- Regeneron Pharmaceuticals, Inc., Tarrytown, NY, 10591, USA. kei.saotome@regeneron.com.
Organizational Affiliation: