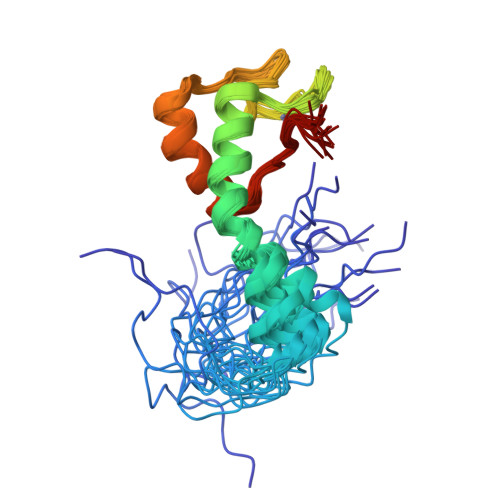
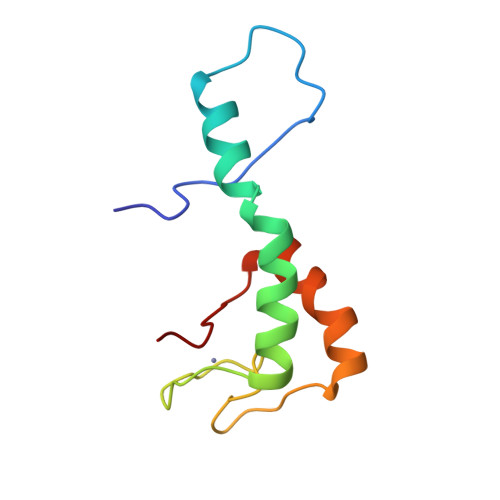
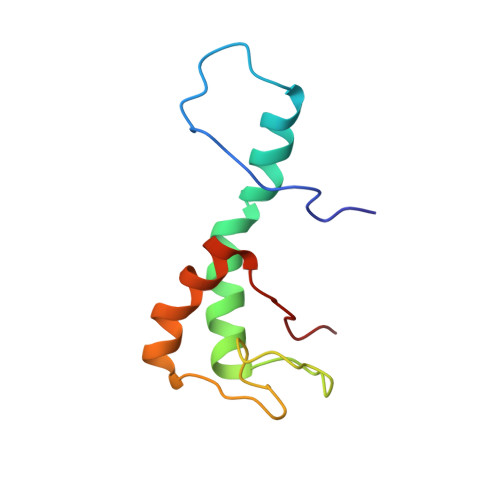
Differences in structure, dynamics, and zinc coordination between isoforms of human ubiquitin ligase UBE3A.
Bregnard, T.A., Fairchild, D., Chen, X., Erlandsen, H., Tarasov, S.G., Walters, K.J., Korzhnev, D.M., Bezsonova, I.(2024) J Biological Chem 301: 108149-108149
- PubMed: 39742997
- DOI: https://doi.org/10.1016/j.jbc.2024.108149
- Primary Citation of Related Structures:
8ENP, 8EPT - PubMed Abstract:
Abnormalities in the expression of the ubiquitin ligase UBE3A (ubiquitin-protein ligase E3A)/E6AP (human papillomavirus E6-associated protein) are implicated in neurological disorders including Angelman syndrome and autism. Human UBE3A is expressed as three protein isoforms that differ in their abundance and subcellular localization. While previous studies indicate isoform-specific functions, the distinct roles of each isoform in human development remain unknown. The isoforms differ only by an extension at the N-terminal end of the AZUL (N-terminal zinc [Zn]-binding domain Amino-terminal Zn finger of the UBE3A Ligase) domain, which tethers UBE3A to the proteasome by interaction with proteasomal subunit Rpn10. Differences in the structure and biophysical properties of UBE3A isoforms likely contribute to their individual functions. Here, we use a combination of NMR spectroscopy and other biophysical and biochemical techniques to identify differences in structure, dynamics, and the Rpn10 binding of the AZUL isoforms. We show that the AZUL domain structure is retained in all three isoforms with an extended N-terminal helix in longer isoforms 2 and 3. Accordingly, all isoforms could effectively associate with the Rpn10. Significant differences between the isoforms were found in their propensities to multimerize where only the longer isoforms 2 and 3 of the AZUL domain could form dimers, which may play a role in the previously observed oligomerization-dependent activation of the UBE3A. Moreover, our NMR relaxation dispersion experiments revealed a dynamic Zn-coordination site in isoforms 1 and 3, but not in isoform 2 of UBE3A, suggesting its possible isoform-specific sensitivity to oxidative stress. This structural and biophysical characterization of the isoforms will advance our understanding of isoform-specific functions of UBE3A and may contribute to future treatment strategies for Angelman syndrome and other UBE3A-related diseases.
Organizational Affiliation:
Department of Molecular Biology and Biophysics, UCONN Health, Farmington, Connecticut, USA.