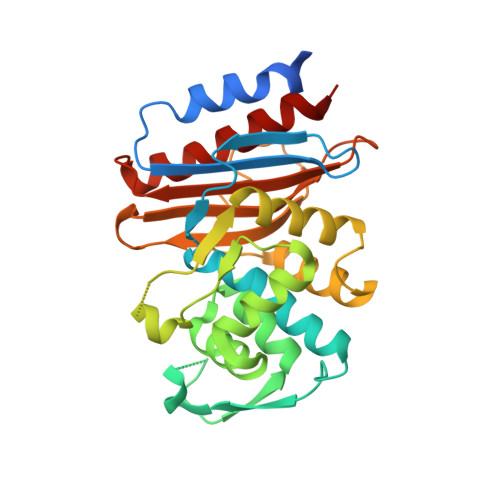
Playing beta-Lactamase Evolution: Metagenomic Class A beta-Lactamase LRA-5 is an Inactive Enzyme Capable of Rendering an Active beta-Lactamase by Introduction of Y69Q and V166E Substitutions
D'Amico Gonzalez, G., Handelsman, J., Centron, D., Gutkind, G., Klinke, S., Power, P.To be published.