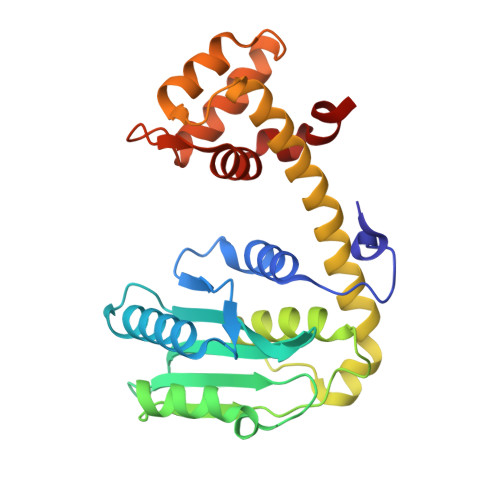
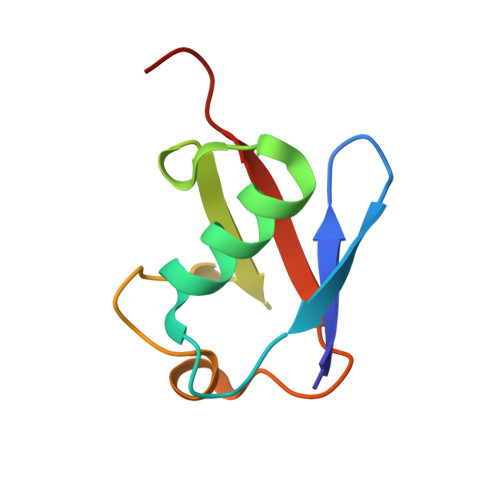
Cocrystallization of ubiquitin-deubiquitinase complexes through disulfide linkage.
Negron Teron, K.I., Das, C.(2023) Acta Crystallogr D Struct Biol 79: 1044-1055
- PubMed: 37877948
- DOI: https://doi.org/10.1107/S2059798323008501
- Primary Citation of Related Structures:
8EFW, 8EFX - PubMed Abstract:
Structural characterization of the recognition of ubiquitin (Ub) by deubiquitinases (DUBs) has largely relied on covalent complexation of the DUB through its catalytic cysteine with a Ub C-terminal electrophile. The Ub electrophiles are accessed through intein chemistry in conjunction with chemical synthesis. Here, it was asked whether DUB-Ub covalent complexes could instead be accessed by simpler disulfide chemistry using a Ub cysteine mutant in which the last glycine has been replaced with a cysteine. The Ub cysteine mutant displayed a wide variability in disulfide formation across a panel of eukaryotic and prokaryotic DUBs, with some showing no detectable reaction while others robustly produced a disulfide complex. Using this approach, two disulfide-linked ubiquitin-bound complexes were crystallized, one involving the Legionella pneumophila effector SdeA DUB and the other involving the Orientia effector OtDUB. These DUBs had previously been crystallized in Ub-bound forms using the C-terminal electrophile strategy and noncovalent complexation, respectively. While the disulfide-linked SdeA DUB-Ub complex crystallized as expected, in the OtDUB complex the disulfide bond to the Ub mutant involved a cysteine that differed from the catalytic cysteine. Disulfide formation with the SdeA DUB catalytic cysteine was accompanied by local distortion of the helix carrying the active-site cysteine, whereas OtDUB reacted with the Ub mutant using a surface-exposed cysteine.
- Department of Chemistry, Purdue University, 560 Oval Drive, West Lafayette, IN 47907, USA.
Organizational Affiliation: