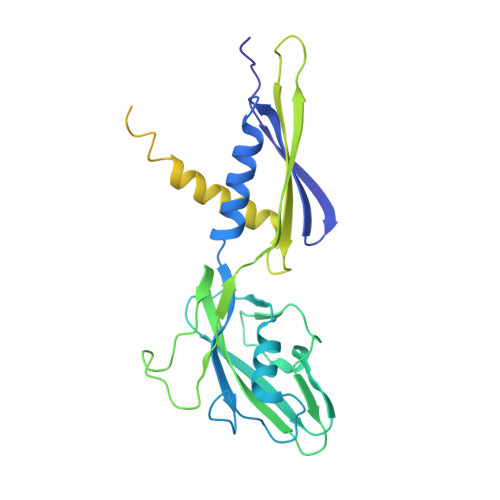
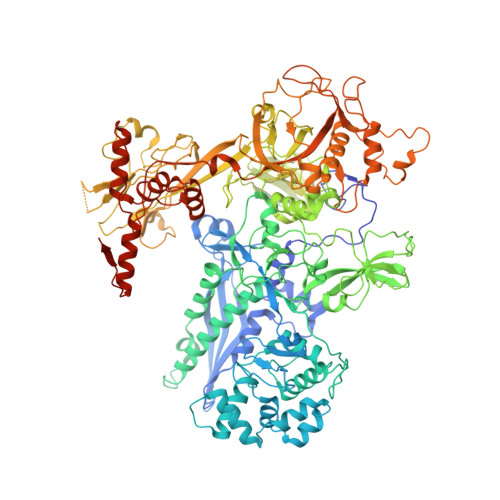
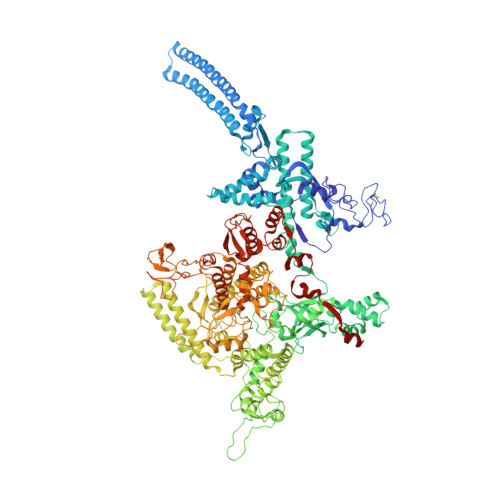
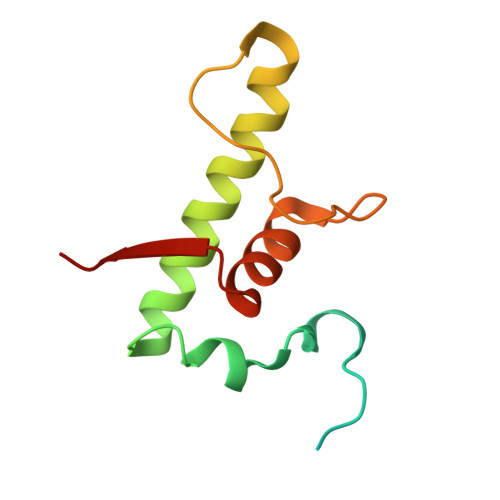
Structural and functional basis of the universal transcription factor NusG pro-pausing activity in Mycobacterium tuberculosis.
Delbeau, M., Omollo, E.O., Froom, R., Koh, S., Mooney, R.A., Lilic, M., Brewer, J.J., Rock, J., Darst, S.A., Campbell, E.A., Landick, R.(2023) Mol Cell 83: 1474-1488.e8
- PubMed: 37116494
- DOI: https://doi.org/10.1016/j.molcel.2023.04.007
- Primary Citation of Related Structures:
8E74, 8E79, 8E82, 8E8M, 8E95 - PubMed Abstract:
Transcriptional pauses mediate regulation of RNA biogenesis. DNA-encoded pause signals trigger pausing by stabilizing RNA polymerase (RNAP) swiveling and inhibiting DNA translocation. The N-terminal domain (NGN) of the only universal transcription factor, NusG/Spt5, modulates pausing through contacts to RNAP and DNA. Pro-pausing NusGs enhance pauses, whereas anti-pausing NusGs suppress pauses. Little is known about pausing and NusG in the human pathogen Mycobacterium tuberculosis (Mtb). We report that MtbNusG is pro-pausing. MtbNusG captures paused, swiveled RNAP by contacts to the RNAP protrusion and nontemplate-DNA wedged between the NGN and RNAP gate loop. In contrast, anti-pausing Escherichia coli (Eco) NGN contacts the MtbRNAP gate loop, inhibiting swiveling and pausing. Using CRISPR-mediated genetics, we show that pro-pausing NGN is required for mycobacterial fitness. Our results define an essential function of mycobacterial NusG and the structural basis of pro- versus anti-pausing NusG activity, with broad implications for the function of all NusG orthologs.
- Laboratory of Molecular Biophysics, The Rockefeller University, New York, NY 10065, USA.
Organizational Affiliation: