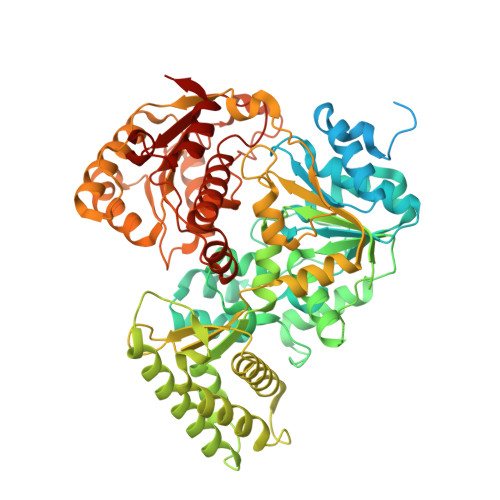
CasDinG is a 5'-3' dsDNA and RNA/DNA helicase with three accessory domains essential for type IV CRISPR immunity.
Domgaard, H., Cahoon, C., Armbrust, M.J., Redman, O., Jolley, A., Thomas, A., Jackson, R.N.(2023) Nucleic Acids Res 51: 8115-8132
- PubMed: 37395408
- DOI: https://doi.org/10.1093/nar/gkad546
- Primary Citation of Related Structures:
8E2W - PubMed Abstract:
CRISPR-associated DinG protein (CasDinG) is essential to type IV-A CRISPR function. Here, we demonstrate that CasDinG from Pseudomonas aeruginosa strain 83 is an ATP-dependent 5'-3' DNA translocase that unwinds double-stranded (ds)DNA and RNA/DNA hybrids. The crystal structure of CasDinG reveals a superfamily 2 helicase core of two RecA-like domains with three accessory domains (N-terminal, arch, and vestigial FeS). To examine the in vivo function of these domains, we identified the preferred PAM sequence for the type IV-A system (5'-GNAWN-3' on the 5'-side of the target) with a plasmid library and performed plasmid clearance assays with domain deletion mutants. Plasmid clearance assays demonstrated that all three domains are essential for type IV-A immunity. Protein expression and biochemical assays suggested the vFeS domain is needed for protein stability and the arch for helicase activity. However, deletion of the N-terminal domain did not impair ATPase, ssDNA binding, or helicase activities, indicating a role distinct from canonical helicase activities that structure prediction tools suggest involves interaction with dsDNA. This work demonstrates CasDinG helicase activity is essential for type IV-A CRISPR immunity as well as the yet undetermined activity of the CasDinG N-terminal domain.
Organizational Affiliation:
Department of Chemistry and Biochemistry, Utah State University, Logan, UT, USA.