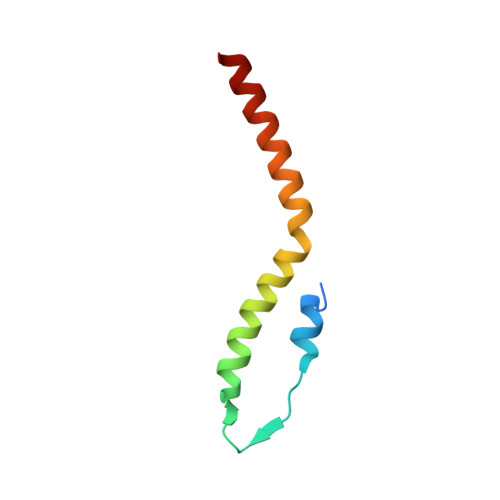
Staphylococcus aureus FtsZ and PBP4 bind to the conformationally dynamic N-terminal domain of GpsB.
Sacco, M.D., Hammond, L.R., Noor, R.E., Bhattacharya, D., McKnight, L.J., Madsen, J.J., Zhang, X., Butler, S.G., Kemp, M.T., Jaskolka-Brown, A.C., Khan, S.J., Gelis, I., Eswara, P., Chen, Y.(2024) Elife 13
- PubMed: 38639993
- DOI: https://doi.org/10.7554/eLife.85579
- Primary Citation of Related Structures:
8E2B, 8E2C - PubMed Abstract:
In the Firmicutes phylum, GpsB is a membrane associated protein that coordinates peptidoglycan synthesis with cell growth and division. Although GpsB has been studied in several bacteria, the structure, function, and interactome of Staphylococcus aureus GpsB is largely uncharacterized. To address this knowledge gap, we solved the crystal structure of the N-terminal domain of S. aureus GpsB, which adopts an atypical, asymmetric dimer, and demonstrates major conformational flexibility that can be mapped to a hinge region formed by a three-residue insertion exclusive to Staphylococci . When this three-residue insertion is excised, its thermal stability increases, and the mutant no longer produces a previously reported lethal phenotype when overexpressed in Bacillus subtilis . In S. aureus , we show that these hinge mutants are less functional and speculate that the conformational flexibility imparted by the hinge region may serve as a dynamic switch to fine-tune the function of the GpsB complex and/or to promote interaction with its various partners. Furthermore, we provide the first biochemical, biophysical, and crystallographic evidence that the N-terminal domain of GpsB binds not only PBP4, but also FtsZ, through a conserved recognition motif located on their C-termini, thus coupling peptidoglycan synthesis to cell division. Taken together, the unique structure of S. aureus GpsB and its direct interaction with FtsZ/PBP4 provide deeper insight into the central role of GpsB in S. aureus cell division.
- Department of Molecular Medicine, Morsani College of Medicine, University of South Florida, Tampa, United States.
Organizational Affiliation: