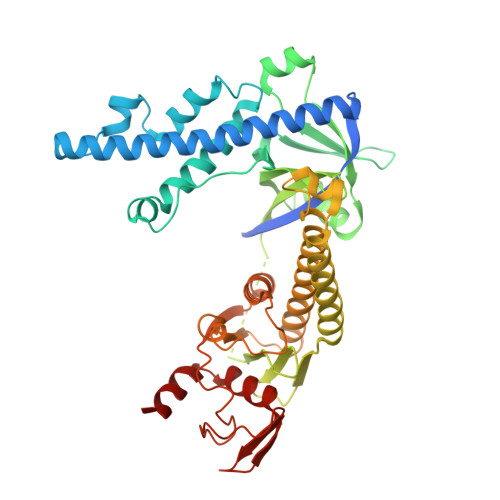
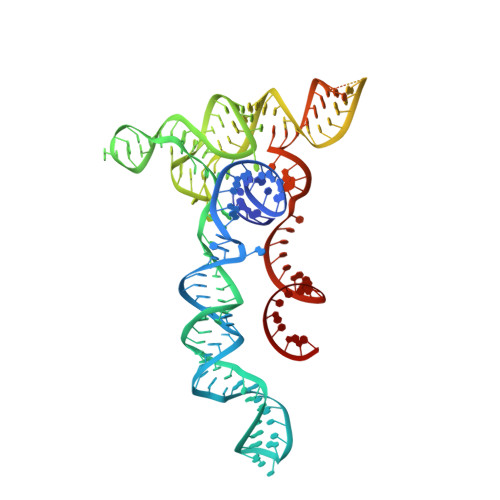
An engineered hypercompact CRISPR-Cas12f system with boosted gene-editing activity.
Wu, T., Liu, C., Zou, S., Lyu, R., Yang, B., Yan, H., Zhao, M., Tang, W.(2023) Nat Chem Biol 19: 1384-1393
- PubMed: 37400536
- DOI: https://doi.org/10.1038/s41589-023-01380-9
- Primary Citation of Related Structures:
8DZJ - PubMed Abstract:
Compact CRISPR-Cas systems offer versatile treatment options for genetic disorders, but their application is often limited by modest gene-editing activity. Here we present enAsCas12f, an engineered RNA-guided DNA endonuclease up to 11.3-fold more potent than its parent protein, AsCas12f, and one-third of the size of SpCas9. enAsCas12f shows higher DNA cleavage activity than wild-type AsCas12f in vitro and functions broadly in human cells, delivering up to 69.8% insertions and deletions at user-specified genomic loci. Minimal off-target editing is observed with enAsCas12f, suggesting that boosted on-target activity does not impair genome-wide specificity. We determine the cryo-electron microscopy (cryo-EM) structure of the AsCas12f-sgRNA-DNA complex at a resolution of 2.9 Å, which reveals dimerization-mediated substrate recognition and cleavage. Structure-guided single guide RNA (sgRNA) engineering leads to sgRNA-v2, which is 33% shorter than the full-length sgRNA, but with on par activity. Together, the engineered hypercompact AsCas12f system enables robust and faithful gene editing in mammalian cells.
- Department of Chemistry, The University of Chicago, Chicago, IL, USA.
Organizational Affiliation: