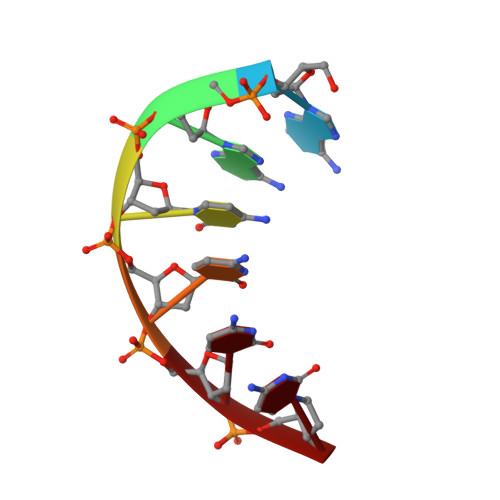
Mapping H + in the Nanoscale (A 2 C 4 ) 2 -Ag 8 Fluorophore.
David, F., Setzler, C., Sorescu, A., Lieberman, R.L., Meilleur, F., Petty, J.T.(2022) J Phys Chem Lett 13: 11317-11322
- PubMed: 36453924
- DOI: https://doi.org/10.1021/acs.jpclett.2c03161
- Primary Citation of Related Structures:
8DYK - PubMed Abstract:
When strands of DNA encapsulate silver clusters, supramolecular optical chromophores develop. However, how a particular structure endows a specific spectrum remains poorly understood. Here, we used neutron diffraction to map protonation in (A 2 C 4 ) 2 -Ag 8 , a green-emitting fluorophore with a "Big Dipper" arrangement of silvers. The DNA host has two substructures with distinct protonation patterns. Three cytosines from each strand collectively chelate handle-like array of three silvers, and calorimetry studies suggest Ag + cross-links. The twisted cytosines are further joined by hydrogen bonds from fully protonated amines. The adenines and their neighboring cytosine from each strand anchor a dipper-like group of five silvers via their deprotonated endo- and exocyclic nitrogens. Typically, exocyclic amines are strongly basic, so their acidification and deprotonation in (A 2 C 4 ) 2 -Ag 8 suggest that silvers perturb the electron distribution in the aromatic nucleobases. The different protonation states in (A 2 C 4 ) 2 -Ag 8 suggest that atomic level structures can pinpoint how to control and tune the electronic spectra of these nanoscale chromophores.
- Department of Chemistry, Furman University, Greenville, South Carolina 29613, United States.
Organizational Affiliation: