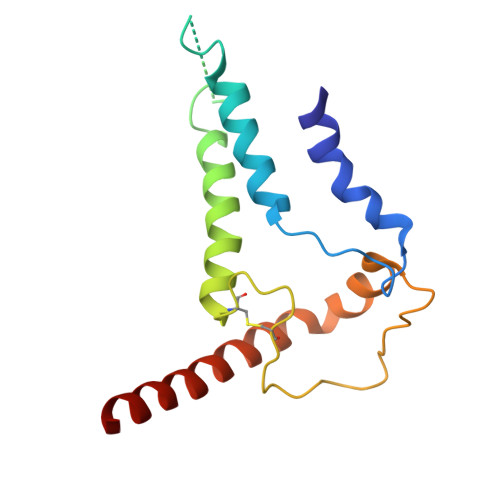
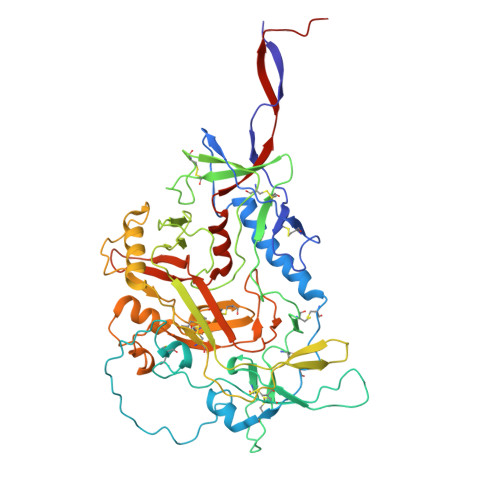
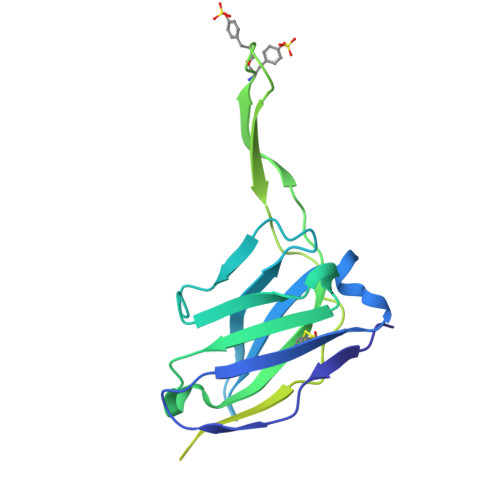
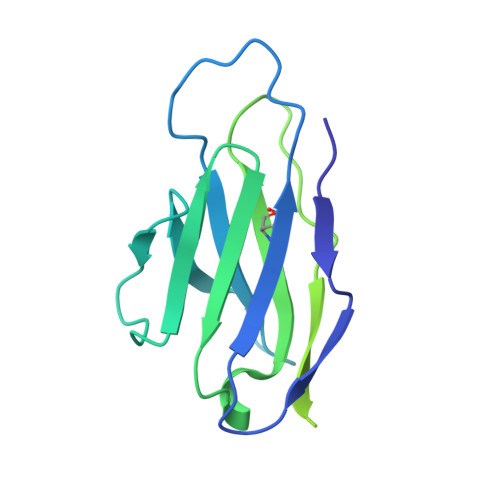
Cryo-EM structures of prefusion SIV envelope trimer.
Gorman, J., Wang, C., Mason, R.D., Nazzari, A.F., Welles, H.C., Zhou, T., Bess Jr., J.W., Bylund, T., Lee, M., Tsybovsky, Y., Verardi, R., Wang, S., Yang, Y., Zhang, B., Rawi, R., Keele, B.F., Lifson, J.D., Liu, J., Roederer, M., Kwong, P.D.(2022) Nat Struct Mol Biol 29: 1080-1091
- PubMed: 36344847
- DOI: https://doi.org/10.1038/s41594-022-00852-1
- Primary Citation of Related Structures:
8DUA, 8DVD - PubMed Abstract:
Simian immunodeficiency viruses (SIVs) are lentiviruses that naturally infect non-human primates of African origin and seeded cross-species transmissions of HIV-1 and HIV-2. Here we report prefusion stabilization and cryo-EM structures of soluble envelope (Env) trimers from rhesus macaque SIV (SIV mac ) in complex with neutralizing antibodies. These structures provide residue-level definition for SIV-specific disulfide-bonded variable loops (V1 and V2), which we used to delineate variable-loop coverage of the Env trimer. The defined variable loops enabled us to investigate assembled Env-glycan shields throughout SIV, which we found to comprise both N- and O-linked glycans, the latter emanating from V1 inserts, which bound the O-link-specific lectin jacalin. We also investigated in situ SIV mac -Env trimers on virions, determining cryo-electron tomography structures at subnanometer resolutions for an antibody-bound complex and a ligand-free state. Collectively, these structures define the prefusion-closed structure of the SIV-Env trimer and delineate variable-loop and glycan-shielding mechanisms of immune evasion conserved throughout SIV evolution.
- Vaccine Research Center, National Institutes of Health, Bethesda, MD, USA.
Organizational Affiliation: