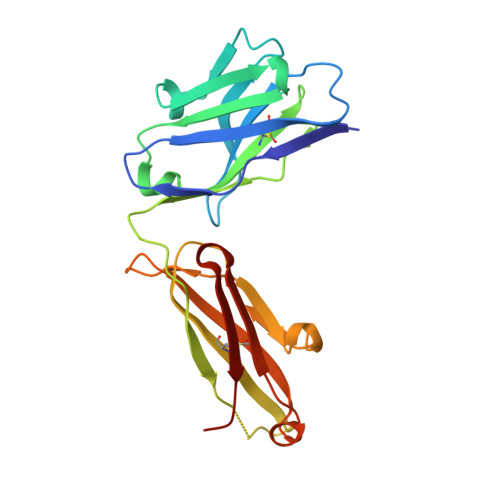
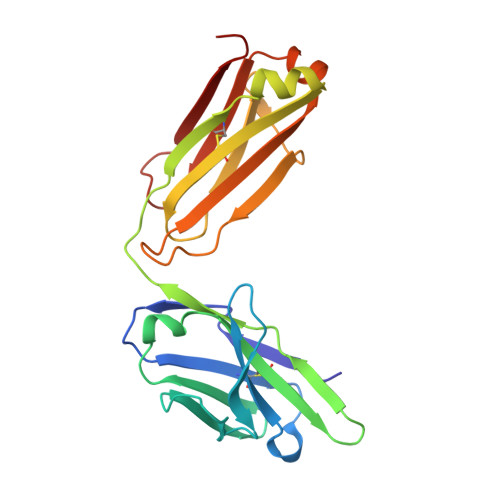
Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Dacon, C., Peng, L., Lin, T.H., Tucker, C., Lee, C.D., Cong, Y., Wang, L., Purser, L., Cooper, A.J.R., Williams, J.K., Pyo, C.W., Yuan, M., Kosik, I., Hu, Z., Zhao, M., Mohan, D., Peterson, M., Skinner, J., Dixit, S., Kollins, E., Huzella, L., Perry, D., Byrum, R., Lembirik, S., Murphy, M., Zhang, Y., Yang, E.S., Chen, M., Leung, K., Weinberg, R.S., Pegu, A., Geraghty, D.E., Davidson, E., Doranz, B.J., Douagi, I., Moir, S., Yewdell, J.W., Schmaljohn, C., Crompton, P.D., Mascola, J.R., Holbrook, M.R., Nemazee, D., Wilson, I.A., Tan, J.(2023) Cell Host Microbe 31: 97
- PubMed: 36347257
- DOI: https://doi.org/10.1016/j.chom.2022.10.010
- Primary Citation of Related Structures:
8DTR, 8DTT, 8DTX - PubMed Abstract:
Humanity has faced three recent outbreaks of novel betacoronaviruses, emphasizing the need to develop approaches that broadly target coronaviruses. Here, we identify 55 monoclonal antibodies from COVID-19 convalescent donors that bind diverse betacoronavirus spike proteins. Most antibodies targeted an S2 epitope that included the K814 residue and were non-neutralizing. However, 11 antibodies targeting the stem helix neutralized betacoronaviruses from different lineages. Eight antibodies in this group, including the six broadest and most potent neutralizers, were encoded by IGHV1-46 and IGKV3-20. Crystal structures of three antibodies of this class at 1.5-1.75-Å resolution revealed a conserved mode of binding. COV89-22 neutralized SARS-CoV-2 variants of concern including Omicron BA.4/5 and limited disease in Syrian hamsters. Collectively, these findings identify a class of IGHV1-46/IGKV3-20 antibodies that broadly neutralize betacoronaviruses by targeting the stem helix but indicate these antibodies constitute a small fraction of the broadly reactive antibody response to betacoronaviruses after SARS-CoV-2 infection.
- Antibody Biology Unit, Laboratory of Immunogenetics, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Rockville, MD 20852, USA.
Organizational Affiliation: