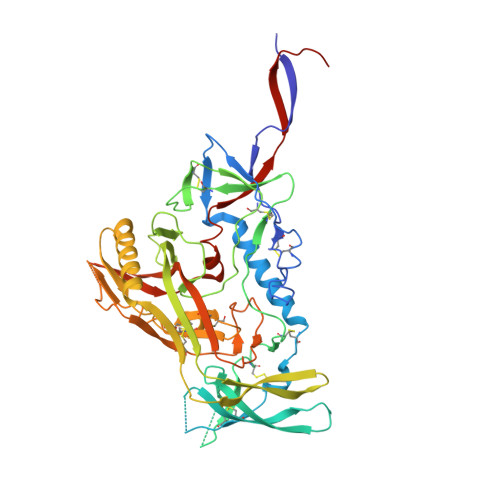
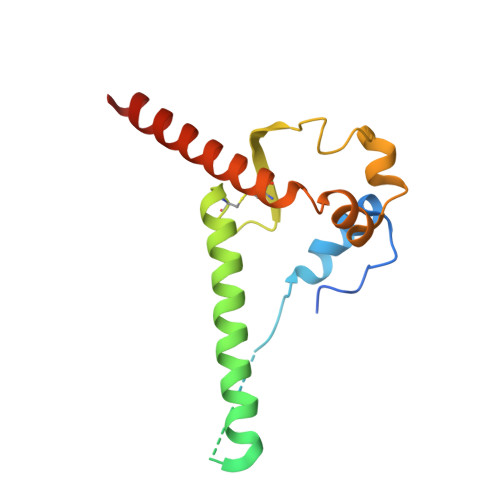
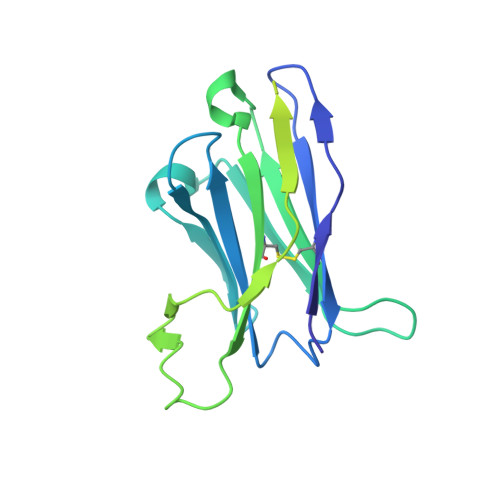
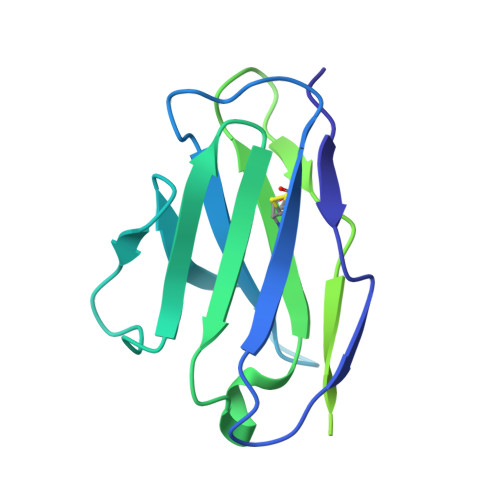
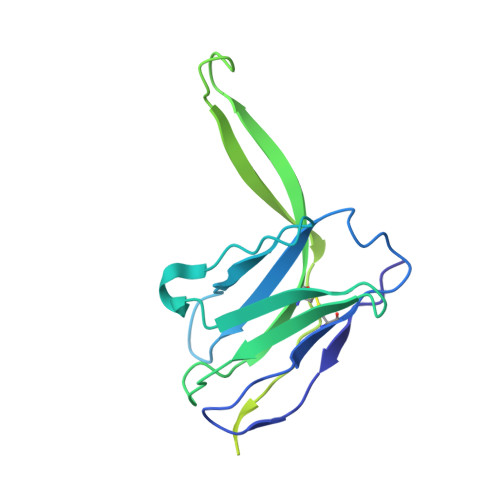
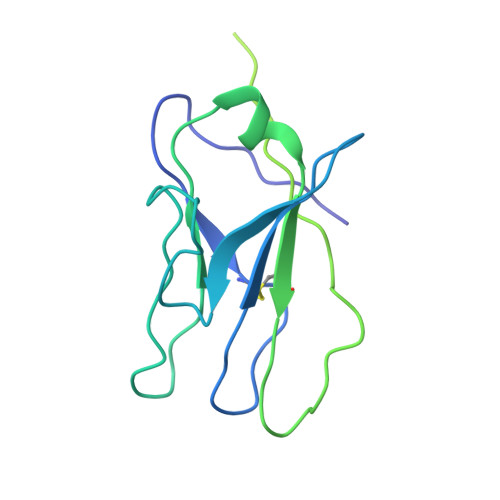
Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir.
Prevost, J., Chen, Y., Zhou, F., Tolbert, W.D., Gasser, R., Medjahed, H., Nayrac, M., Nguyen, D.N., Gottumukkala, S., Hessell, A.J., Rao, V.B., Pozharski, E., Huang, R.K., Matthies, D., Finzi, A., Pazgier, M.(2023) Nat Commun 14: 6710-6710
- PubMed: 37872202
- DOI: https://doi.org/10.1038/s41467-023-42500-2
- Primary Citation of Related Structures:
8CZZ, 8DOK, 8G6U, 8TTW - PubMed Abstract:
The HIV-1 entry inhibitor temsavir prevents the viral receptor CD4 (cluster of differentiation 4) from interacting with the envelope glycoprotein (Env) and blocks its conformational changes. To do this, temsavir relies on the presence of a residue with small side chain at position 375 in Env and is unable to neutralize viral strains like CRF01_AE carrying His375. Here we investigate the mechanism of temsavir resistance and show that residue 375 is not the sole determinant of resistance. At least six additional residues within the gp120 inner domain layers, including five distant from the drug-binding pocket, contribute to resistance. A detailed structure-function analysis using engineered viruses and soluble trimer variants reveals that the molecular basis of resistance is mediated by crosstalk between His375 and the inner domain layers. Furthermore, our data confirm that temsavir can adjust its binding mode to accommodate changes in Env conformation, a property that likely contributes to its broad antiviral activity.
- Centre de Recherche du CHUM, Montreal, QC, Canada.
Organizational Affiliation: