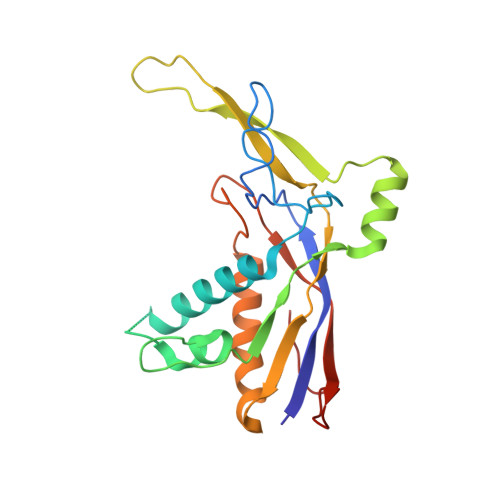
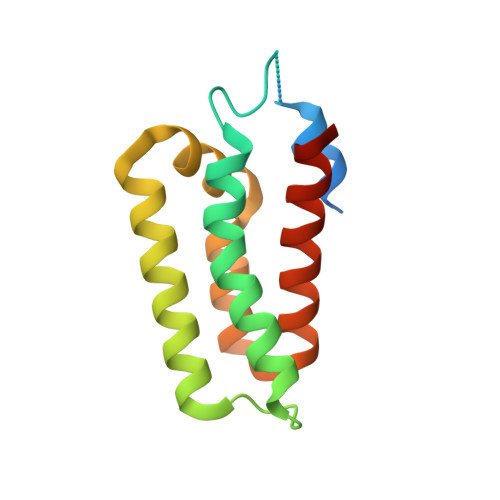
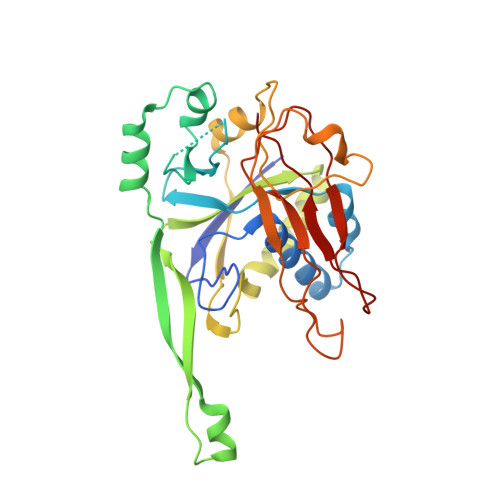
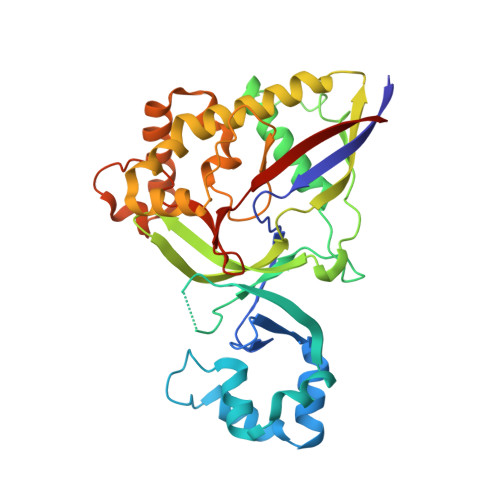
The structure of a Type III-A CRISPR-Cas effector complex reveals conserved and idiosyncratic contacts to target RNA and crRNA among Type III-A systems.
Paraan, M., Nasef, M., Chou-Zheng, L., Khweis, S.A., Schoeffler, A.J., Hatoum-Aslan, A., Stagg, S.M., Dunkle, J.A.(2023) PLoS One 18: e0287461-e0287461
- PubMed: 37352230
- DOI: https://doi.org/10.1371/journal.pone.0287461
- Primary Citation of Related Structures:
8DO6 - PubMed Abstract:
Type III CRISPR-Cas systems employ multiprotein effector complexes bound to small CRISPR RNAs (crRNAs) to detect foreign RNA transcripts and elicit a complex immune response that leads to the destruction of invading RNA and DNA. Type III systems are among the most widespread in nature, and emerging interest in harnessing these systems for biotechnology applications highlights the need for detailed structural analyses of representatives from diverse organisms. We performed cryo-EM reconstructions of the Type III-A Cas10-Csm effector complex from S. epidermidis bound to an intact, cognate target RNA and identified two oligomeric states, a 276 kDa complex and a 318 kDa complex. 3.1 Å density for the well-ordered 276 kDa complex allowed construction of atomic models for the Csm2, Csm3, Csm4 and Csm5 subunits within the complex along with the crRNA and target RNA. We also collected small-angle X-ray scattering data which was consistent with the 276 kDa Cas10-Csm architecture we identified. Detailed comparisons between the S. epidermidis Cas10-Csm structure and the well-resolved bacterial (S. thermophilus) and archaeal (T. onnurineus) Cas10-Csm structures reveal differences in how the complexes interact with target RNA and crRNA which are likely to have functional ramifications. These structural comparisons shed light on the unique features of Type III-A systems from diverse organisms and will assist in improving biotechnologies derived from Type III-A effector complexes.
- National Center for In-situ Tomographic Ultramicroscopy, Simons Electron Microscopy Center, New York Structural Biology Center, New York, NY, United States of America.
Organizational Affiliation: