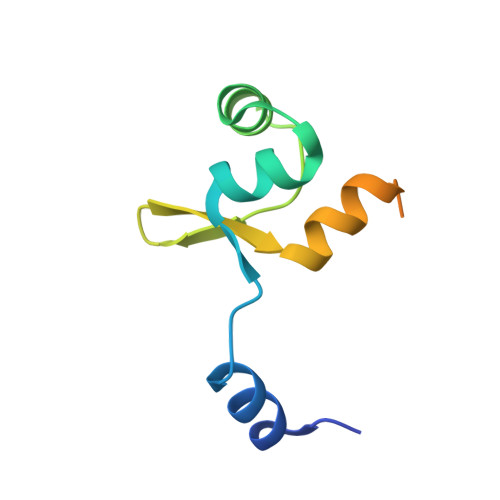
Crystallographic and X-ray scattering study of RdfS, a recombination directionality factor from an integrative and conjugative element.
Verdonk, C.J., Marshall, A.C., Ramsay, J.P., Bond, C.S.(2022) Acta Crystallogr D Struct Biol 78: 1210-1220
- PubMed: 36189741
- DOI: https://doi.org/10.1107/S2059798322008579
- Primary Citation of Related Structures:
8DGL - PubMed Abstract:
The recombination directionality factors from Mesorhizobium spp. (RdfS) are involved in regulating the excision and transfer of integrative and conjugative elements. Here, solution small-angle X-ray scattering, and crystallization and preliminary structure solution of RdfS from Mesorhizobium japonicum R7A are presented. RdfS crystallizes in space group P2 1 2 1 2 1 , with evidence of eightfold rotational crystallographic/noncrystallographic symmetry. Initial structure determination by molecular replacement using ab initio models yielded a partial model (three molecules), which was completed after manual inspection revealed unmodelled electron density. The finalized crystal structure of RdfS reveals a head-to-tail polymer forming left-handed superhelices with large solvent channels. Additionally, RdfS has significant disorder in the C-terminal region of the protein, which is supported by the solution scattering data and the crystal structure. The steps taken to finalize structure determination, as well as the scattering and crystallographic characteristics of RdfS, are discussed.
Organizational Affiliation:
School of Molecular Sciences, University of Western Australia, Perth, Western Australia 6009, Australia.