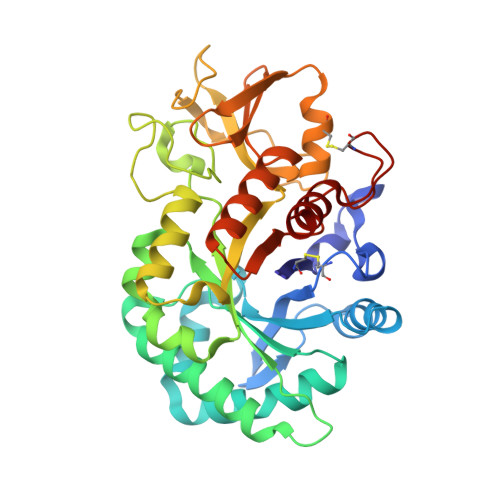
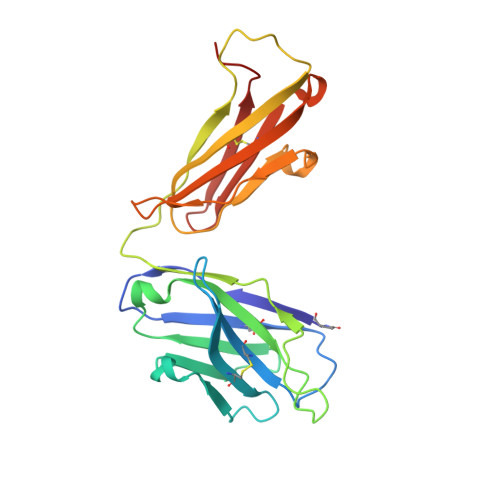
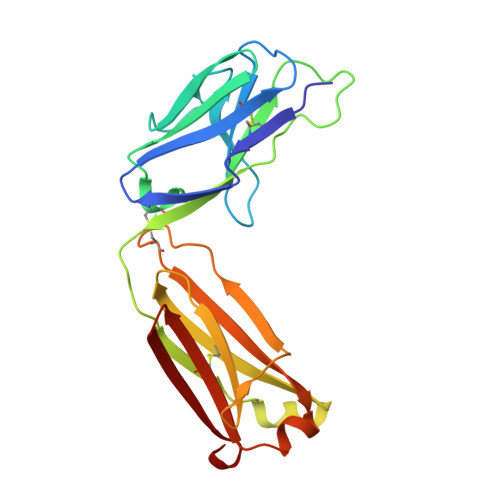
A novel humanized Chi3l1 blocking antibody attenuates acetaminophen-induced liver injury in mice.
Li, L., Wen, Y., Wrapp, D., Jeong, J., Zhao, P., Xiong, W., Atkins, C.L., Shan, Z., Hui, D., McLellan, J.S., Zhang, N., Ju, C., An, Z.(2023) Antib Ther 6: 1-12
- PubMed: 36683763
- DOI: https://doi.org/10.1093/abt/tbac027
- Primary Citation of Related Structures:
8DF1 - PubMed Abstract:
Acetaminophen (APAP) overdose is a leading cause of acute liver injury in the USA. The chitinase 3-like-1 (Chi3l1) protein contributes to APAP-induced liver injury (AILI) by promoting hepatic platelet recruitment. Here, we report the development of a Chi3l1-targeting antibody as a potential therapy for AILI. By immunizing a rabbit successively with the human and mouse Chi3l1 proteins, we isolated cross-reactive monoclonal antibodies (mAbs) from single memory B cells. One of the human and mouse Chi3l1 cross-reactive mAbs was humanized and characterized in both in vitro and in vivo biophysical and biological assays. X-ray crystallographic analysis of the lead antibody C59 in complex with the human Chi3l1 protein revealed that the kappa light contributes to majority of the antibody-antigen interaction; and that C59 binds to the 4α-5β loop and 4α-helix of Chi3l1, which is a functional epitope and hotspot for the development of Chi3l1 blocking antibodies. We humanized the C59 antibody by complementarity-determining region grafting and kappa chain framework region reverse mutations. The humanized C59 antibody exhibited similar efficacy as the parental rabbit antibody C59 in attenuating AILI in vivo . Our findings validate Chi3l1 as a potential drug target for AILI and provide proof of concept of developing Chi3l1 blocking antibody as a therapy for the treatment of AILI.
- Texas Therapeutics Institute, Brown Foundation Institute of Molecular Medicine, McGovern Medical School, The University of Texas Health Science Center at Houston, Houston, TX 77030, USA.
Organizational Affiliation: