Native serotonin transporter in complex with 15B8 Fab in the presence of methamphetamine
Yang, D., Gouaux, E.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Transporter | 539 | Sus scrofa | Mutation(s): 0 Membrane Entity: Yes | 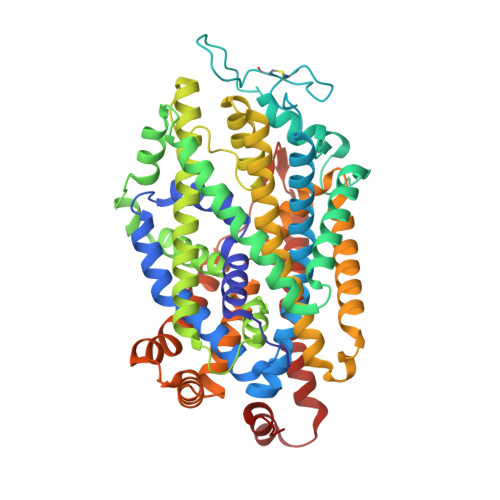 | |
UniProt | |||||
Find proteins for F1RN70 (Sus scrofa) Explore F1RN70 Go to UniProtKB: F1RN70 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | F1RN70 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 15B8 Fab heavy chain variable domain | 118 | Mus musculus | Mutation(s): 0 | 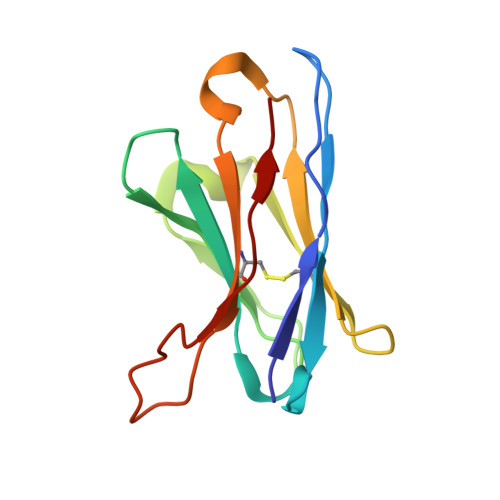 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 15B8 Fab light chain variable domain | 110 | Mus musculus | Mutation(s): 0 | 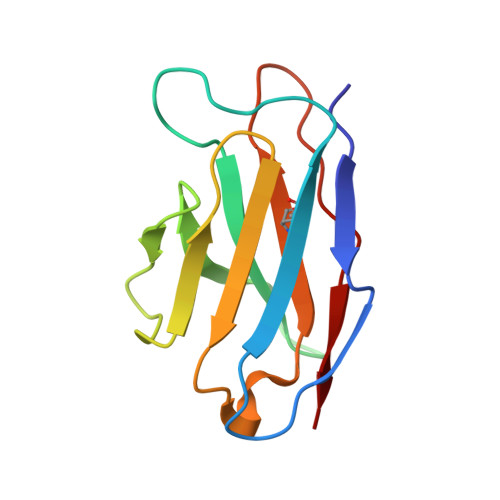 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| LMT (Subject of Investigation/LOI) Query on LMT | K [auth A] | DODECYL-BETA-D-MALTOSIDE C24 H46 O11 NLEBIOOXCVAHBD-QKMCSOCLSA-N |  | ||
| Y01 Query on Y01 | I [auth A], J [auth A] | CHOLESTEROL HEMISUCCINATE C31 H50 O4 WLNARFZDISHUGS-MIXBDBMTSA-N |  | ||
| B40 (Subject of Investigation/LOI) Query on B40 | H [auth A] | (2S)-N-methyl-1-phenylpropan-2-amine C10 H15 N MYWUZJCMWCOHBA-VIFPVBQESA-N |  | ||
| CL Query on CL | E [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| NA Query on NA | F [auth A], G [auth A] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Howard Hughes Medical Institute (HHMI) | United States | -- |
| National Institutes of Health/National Center for Research Resources (NIH/NCRR) | United States | -- |