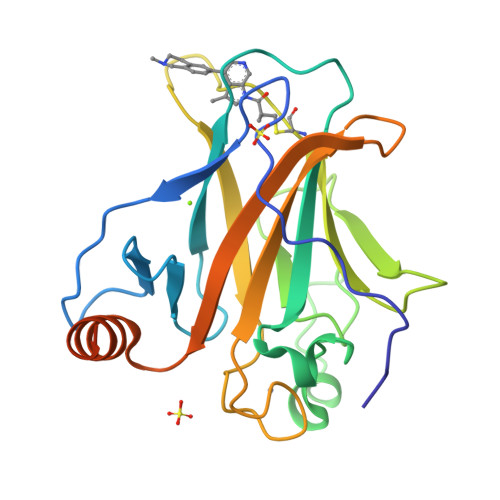
A Small Molecule Reacts with the p53 Somatic Mutant Y220C to Rescue Wild-type Thermal Stability.
Guiley, K.Z., Shokat, K.M.(2023) Cancer Discov 13: 56-69
- PubMed: 36197521
- DOI: https://doi.org/10.1158/2159-8290.CD-22-0381
- Primary Citation of Related Structures:
8DC4, 8DC6, 8DC7, 8DC8 - PubMed Abstract:
The transcription factor and tumor suppressor protein p53 is the most frequently mutated and inactivated gene in cancer. Mutations in p53 result in deregulated cell proliferation and genomic instability, both hallmarks of cancer. There are currently no therapies available that directly target mutant p53 to rescue wild-type function. In this study, we identify covalent compsounds that selectively react with the p53 somatic mutant cysteine Y220C and restore wild-type thermal stability. The tumor suppressor p53 is the most mutated gene in cancer, and yet no therapeutics to date directly target the mutated protein to rescue wild-type function. In this study, we identify the first allele-specific compound that selectively reacts with the cysteine p53 Y220C to rescue wild-type thermal stability and gene activation. See related commentary by Lane and Verma, p. 14. This article is highlighted in the In This Issue feature, p. 1.
Organizational Affiliation:
Department of Cellular and Molecular Pharmacology and Howard Hughes Medical Institute, University of California, San Francisco, San Francisco, California.