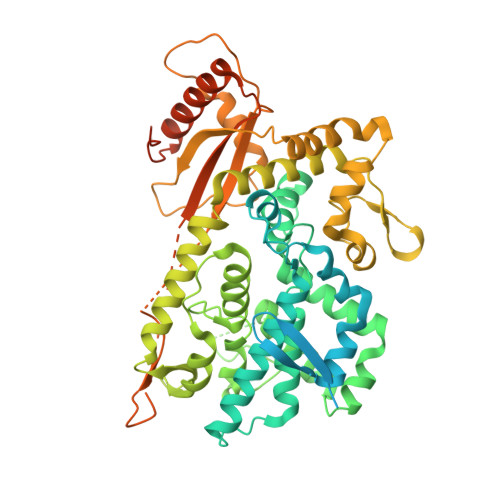
Protein oxidation increases SAMHD1 binding ssDNA via its regulatory site.
Simermeyer, T.L., Batalis, S., Rogers, L.C., Zalesak, O.J., Hollis, T.(2023) Nucleic Acids Res 51: 7014-7024
- PubMed: 37246644
- DOI: https://doi.org/10.1093/nar/gkad447
- Primary Citation of Related Structures:
8D9J - PubMed Abstract:
SAMHD1 dNTP hydrolase activity places it at the crossroad of several important biological pathways, such as viral restriction, cell cycle regulation, and innate immunity. Recently, a dNTPase independent function for SAMHD1 in homologous recombination (HR) of DNA double-strand breaks has been identified. SAMHD1 function and activity is regulated by several post-translational modifications, including protein oxidation. Here, we showed that oxidation of SAMHD1 increases ssDNA binding affinity and occurs in a cell cycle-dependent manner during S phase consistent with a role in HR. We determined the structure of oxidized SAMHD1 in complex with ssDNA. The enzyme binds ssDNA at the regulatory sites at the dimer interface. We propose a mechanism that oxidation of SAMHD1 acts as a functional switch to toggle between dNTPase activity and DNA binding.
Organizational Affiliation:
Department of Biochemistry, Wake Forest School of Medicine, Winston-Salem, NC, USA.