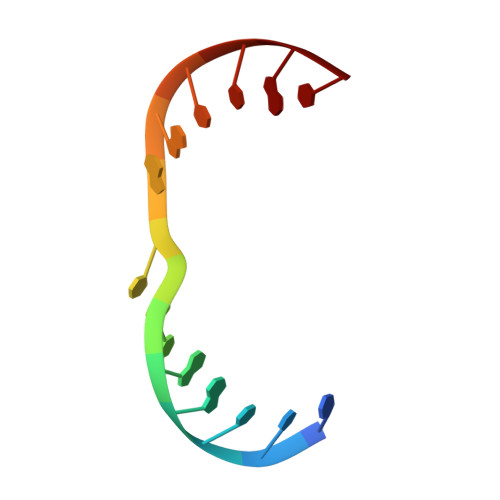The Rule of Thirds: Controlling Junction Chirality and Polarity in 3D DNA Tiles.
Vecchioni, S., Lu, B., Janowski, J., Woloszyn, K., Jonoska, N., Seeman, N.C., Mao, C., Ohayon, Y.P., Sha, R.(2023) Small 19: e2206511-e2206511
- PubMed: 36585389
- DOI: https://doi.org/10.1002/smll.202206511
- Primary Citation of Related Structures:
7SPL, 8D93 - PubMed Abstract:
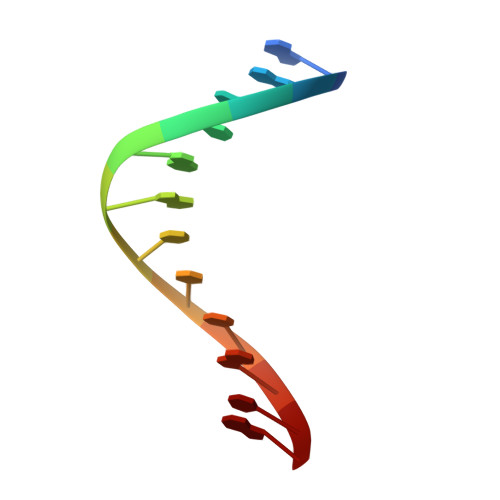
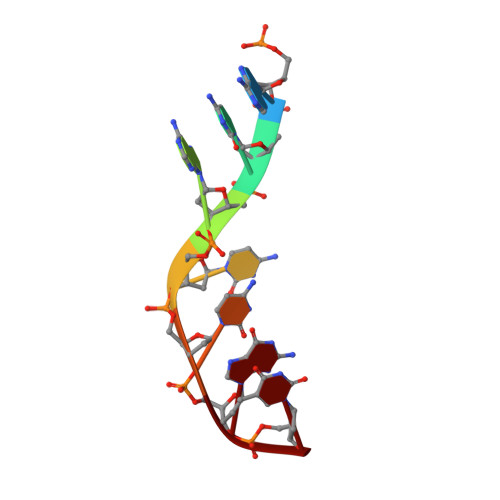
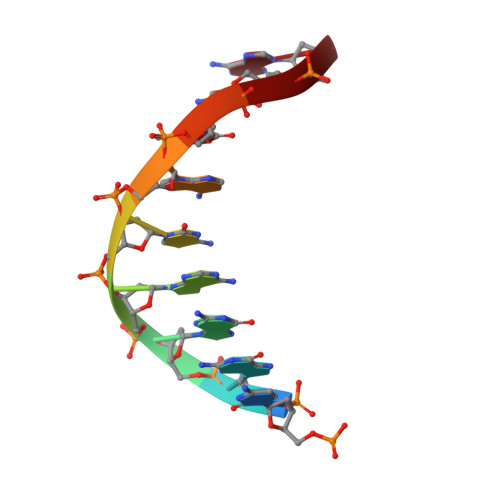
The successful self-assembly of tensegrity triangle DNA crystals heralded the ability to programmably construct macroscopic crystalline nanomaterials from rationally-designed, nanoscale components. This 3D DNA tile owes its "tensegrity" nature to its three rotationally stacked double helices locked together by the tensile winding of a center strand segmented into 7 base pair (bp) inter-junction regions, corresponding to two-thirds of a helical turn of DNA. All reported tensegrity triangles to date have employed
Z + 2 / 3 ) \[\left( {Z{\bm{ + }}2{\bf /}3} \right)\] turn inter-junction segments, yielding right-handed, antiparallel, "J1" junctions. Here a minimal DNA triangle motif consisting of 3-bp inter-junction segments, or one-third of a helical turn is reported. It is found that the minimal motif exhibits a reversed morphology with a left-handed tertiary structure mediated by a locally-parallel Holliday junction-the "L1" junction. This parallel junction yields a predicted helical groove matching pattern that breaks the pseudosymmetry between tile faces, and the junction morphology further suggests a folding mechanism. A Rule of Thirds by which supramolecular chirality can be programmed through inter-junction DNA segment length is identified. These results underscore the role that global topological forces play in determining local DNA architecture and ultimately point to an under-explored class of self-assembling, chiral nanomaterials for topological processes in biological systems. - Department of Chemistry, New York University, New York, NY, 10003, USA.
Organizational Affiliation: