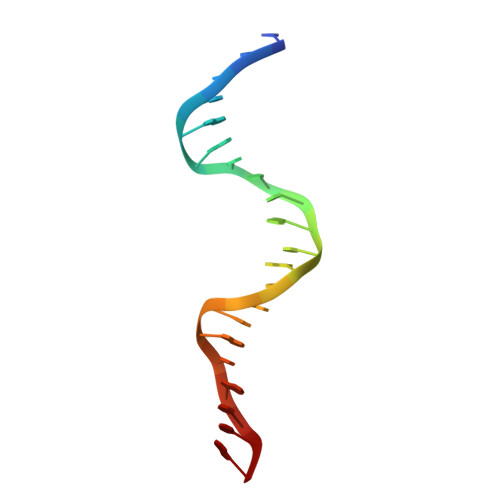
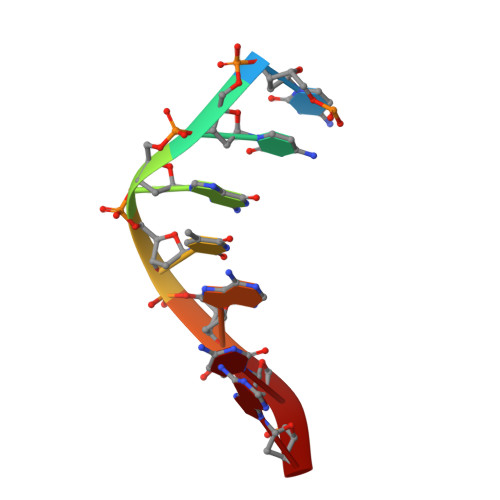
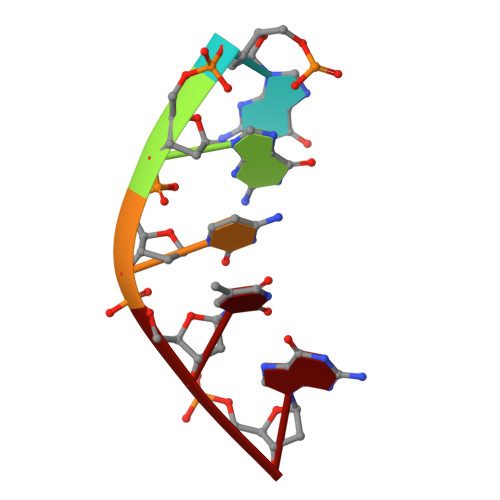
Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Lu, B., Woloszyn, K., Ohayon, Y.P., Yang, B., Zhang, C., Mao, C., Seeman, N.C., Vecchioni, S., Sha, R.(2023) Angew Chem Int Ed Engl 62: e202213451-e202213451
- PubMed: 36520622
- DOI: https://doi.org/10.1002/anie.202213451
- Primary Citation of Related Structures:
8CS1, 8CS2, 8CS3, 8CS4, 8CS5, 8CS6, 8CS7, 8CS8, 8CYM, 8CYN, 8DAG, 8DAH, 8DCJ - PubMed Abstract:
Non-canonical interactions in DNA remain under-explored in DNA nanotechnology. Recently, many structures with non-canonical motifs have been discovered, notably a hexagonal arrangement of typically rhombohedral DNA tensegrity triangles that forms through non-canonical sticky end interactions. Here, we find a series of mechanisms to program a hexagonal arrangement using: the sticky end sequence; triangle edge torsional stress; and crystallization condition. We showcase cross-talking between Watson-Crick and non-canonical sticky ends in which the ratio between the two dictates segregation by crystal forms or combination into composite crystals. Finally, we develop a method for reconfiguring the long-range geometry of formed crystals from rhombohedral to hexagonal and vice versa. These data demonstrate fine control over non-canonical motifs and their topological self-assembly. This will vastly increase the programmability, functionality, and versatility of rationally designed DNA constructs.
- Department of Chemistry, New York University, New York, NY 10003, USA.
Organizational Affiliation: