Novel druggable space in human KRAS G13D discovered using structural bioinformatics and a P-loop targeting monoclonal antibody.
Jungholm, O., Trkulja, C., Moche, M., Srinivasa, S.P., Christakopoulou, M.N., Davidson, M., Reymer, A., Jardemark, K., Fogaca, R.L., Ashok, A., Jeffries, G., Ampah-Korsah, H., Strandback, E., Andrell, J., Nyman, T., Nouairia, G., Orwar, O.(2024) Sci Rep 14: 19656-19656
- PubMed: 39179604
- DOI: https://doi.org/10.1038/s41598-024-70217-9
- Primary Citation of Related Structures:
8BLR, 8CPR - PubMed Abstract:
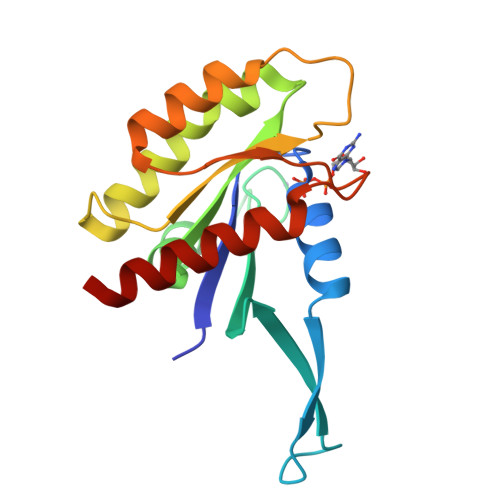
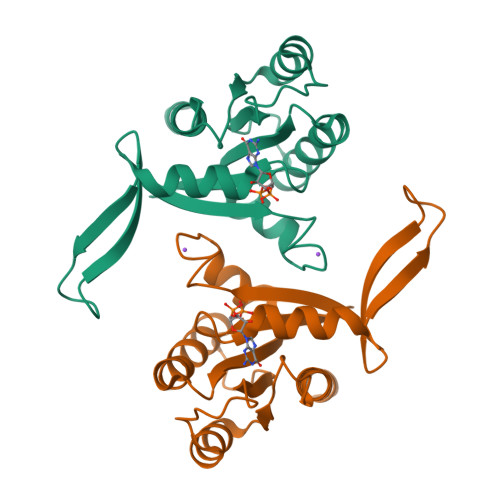
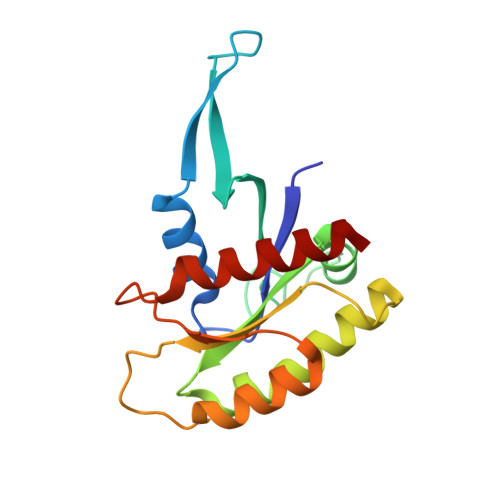
KRAS belongs to a family of small GTPases that act as binary switches upstream of several signalling cascades, controlling proliferation and survival of cells. Mutations in KRAS drive oncogenesis, especially in pancreatic, lung, and colorectal cancers (CRC). Although historic attempts at targeting mutant KRAS with small molecule inhibitors have proven challenging, there are recent successes with the G12C, and G12D mutations. However, clinically important RAS mutations such as G12V, G13D, Q61L, and A146T, remain elusive drug targets, and insights to their structural landscape is of critical importance to develop novel, and effective therapeutic concepts. We present a fully open, P-loop exposing conformer of KRAS G13D by X-ray crystallography at 1.4-2.4 Å resolution in Mg 2+ -free phosphate and malonate buffers. The G13D conformer has the switch-I region displaced in an upright position leaving the catalytic core fully exposed. To prove that this state is druggable, we developed a P-loop-targeting monoclonal antibody (mAb). The mAb displayed high-affinity binding to G13D and was shown using high resolution fluorescence microscopy to be spontaneously taken up by G13D-mutated HCT 116 cells (human CRC derived) by macropinocytosis. The mAb inhibited KRAS signalling in phosphoproteomic and genomic studies. Taken together, the data propose novel druggable space of G13D that is reachable in the cellular context. It is our hope that these findings will stimulate attempts to drug this fully open state G13D conformer using mAbs or other modalities.
Organizational Affiliation:
Department of Physiology and Pharmacology, Karolinska Institutet, 171 77, Stockholm, Sweden.