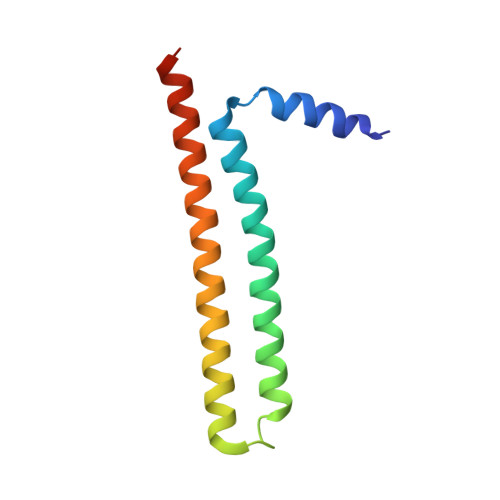
Structural analysis of ING3 protein and histone H3 binding.
Ferreras-Gutierrez, M., Chaves-Arquero, B., Gonzalez-Magana, A., Merino, N., Amusategui-Mateu, I., Huecas, S., Medrano, F.J., Blanco, F.J.(2023) Int J Biol Macromol 242: 124724-124724
- PubMed: 37148949
- DOI: https://doi.org/10.1016/j.ijbiomac.2023.124724
- Primary Citation of Related Structures:
7ZMX, 8COK - PubMed Abstract:
Proteins belonging to the ING family regulate the transcriptional state of chromatin by recruiting remodeling complexes to sites with histone H3 trimethylated at Lysine 4 (H3K4me3). This modification is recognized by the Plant HomeoDomain (PHD) present at the C-terminal region of the five ING proteins. ING3 facilitates acetylation of histones H2A and H4 by the NuA4-Tip60 MYST histone acetyl transferase complex, and it has been proposed to be an oncoprotein. The crystal structure of the N-terminal domain of ING3 shows that it forms homodimers with an antiparallel coiled-coil fold. The crystal structure of the PHD is similar to those of its four homologs. These structures explain the possible deleterious effects of ING3 mutations detected in tumors. The PHD binds histone H3K4me3 with low-micromolar, and binds the non-methylated histone with a 54-fold reduced affinity. Our structure explains the impact of site directed mutagenesis experiments on histone recognition. These structural features could not be confirmed for the full-length protein as solubility was insufficient for structural studies, but the structure of its folded domains suggest a conserved structural organization for the ING proteins as homodimers and bivalent readers of the histone H3K4me3 mark.
- Centro de Investigaciones Biológicas Margarita Salas (CIB), CSIC, Madrid 28040, Spain.
Organizational Affiliation: