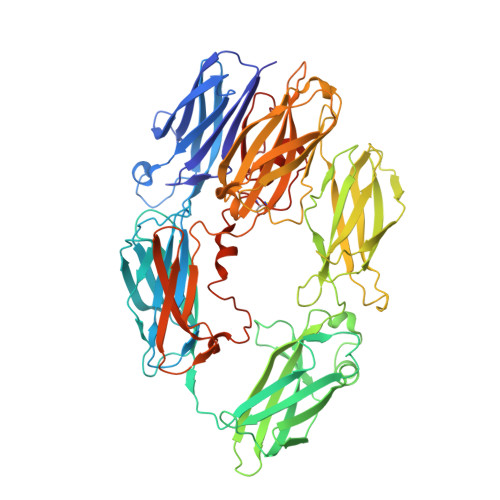
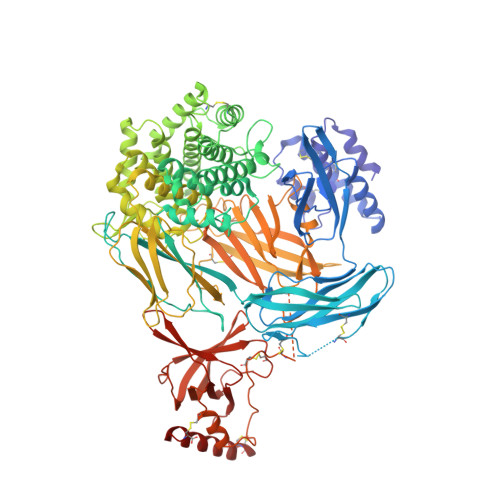
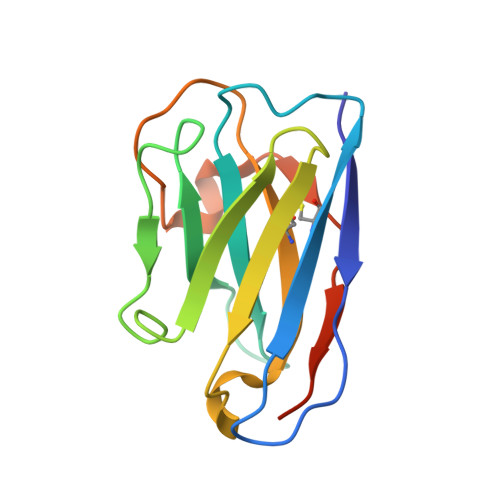
Characterization of the bispecific VHH antibody gefurulimab (ALXN1720) targeting complement component 5, and designed for low volume subcutaneous administration.
Jindal, S., Pedersen, D.V., Gera, N., Chandler, J., Patel, R., Neill, A., Cone, J., Zhang, Y., Yuan, C.X., Millman, E.E., Carlin, D., Puffer, B., Sheridan, D., Andersen, G.R., Tamburini, P.(2023) Mol Immunol 165: 29-41
- PubMed: 38142486
- DOI: https://doi.org/10.1016/j.molimm.2023.12.004
- Primary Citation of Related Structures:
8COE, 8COH - PubMed Abstract:
The bispecific antibody gefurulimab (also known as ALXN1720) was developed to provide patients with a subcutaneous treatment option for chronic disorders involving activation of the terminal complement pathway. Gefurulimab blocks the enzymatic cleavage of complement component 5 (C5) into the biologically active C5a and C5b fragments, which triggers activation of the terminal complement cascade. Heavy-chain variable region antigen-binding fragment (VHH) antibodies targeting C5 and human serum albumin (HSA) were isolated from llama immune-based libraries and humanized. Gefurulimab comprises an N-terminal albumin-binding VHH connected to a C-terminal C5-binding VHH via a flexible linker. The purified bispecific VHH antibody has the expected exact size by mass spectrometry and can be formulated at greater than 100 mg/mL. Gefurulimab binds tightly to human C5 and HSA with dissociation rate constants at pH 7.4 of 54 pM and 0.9 nM, respectively, and cross-reacts with C5 and serum albumin from cynomolgus monkeys. Gefurulimab can associate with C5 and albumin simultaneously, and potently inhibits the terminal complement activity from human serum initiated by any of the three complement pathways in Wieslab assays. Electron microscopy and X-ray crystallography revealed that the isolated C5-binding VHH recognizes the macroglobulin (MG) 4 and MG5 domains of the antigen and thereby is suggested to sterically prevent C5 binding to its activating convertase. Gefurulimab also inhibits complement activity supported by the rare C5 allelic variant featuring an R885H substitution in the MG7 domain. Taken together, these data suggest that gefurulimab may be a promising candidate for the potential treatment of complement-mediated disorders.
- Alexion, AstraZeneca Rare Disease, 100 College Street, New Haven, CT 06510, USA.
Organizational Affiliation: