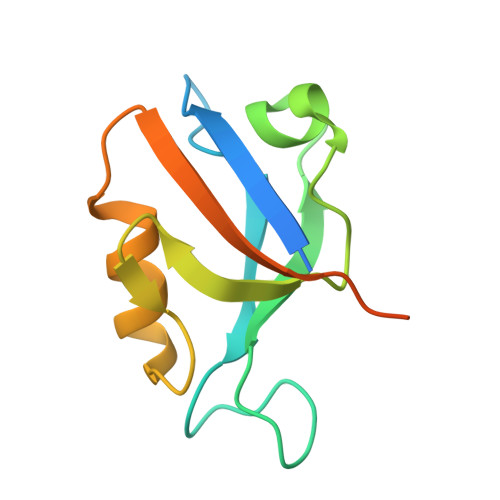
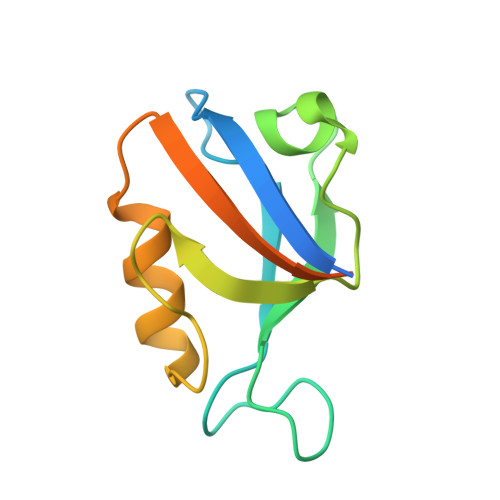
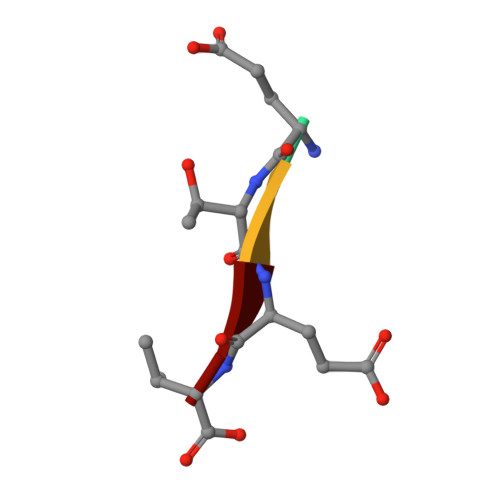
Identification of small molecule antivirals against HTLV-1 by targeting the hDLG1-Tax-1 protein-protein interaction.
Maseko, S.B., Brammerloo, Y., Van Molle, I., Sogues, A., Martin, C., Gorgulla, C., Plant, E., Olivet, J., Blavier, J., Ntombela, T., Delvigne, F., Arthanari, H., El Hajj, H., Bazarbachi, A., Van Lint, C., Salehi-Ashtiani, K., Remaut, H., Ballet, S., Volkov, A.N., Twizere, J.C.(2023) Antiviral Res 217: 105675-105675
- PubMed: 37481039
- DOI: https://doi.org/10.1016/j.antiviral.2023.105675
- Primary Citation of Related Structures:
8CN1, 8CN3 - PubMed Abstract:
Human T-cell leukemia virus type-1 (HTLV-1) is the first pathogenic retrovirus discovered in human. Although HTLV-1-induced diseases are well-characterized and linked to the encoded Tax-1 oncoprotein, there is currently no strategy to target Tax-1 functions with small molecules. Here, we analyzed the binding of Tax-1 to the human homolog of the drosophila discs large tumor suppressor (hDLG1/SAP97), a multi-domain scaffolding protein involved in Tax-1-transformation ability. We have solved the structures of the PDZ binding motif (PBM) of Tax-1 in complex with the PDZ1 and PDZ2 domains of hDLG1 and assessed the binding of 10 million molecules by virtual screening. Among the 19 experimentally confirmed compounds, one systematically inhibited the Tax-1-hDLG1 interaction in different biophysical and cellular assays, as well as HTLV-1 cell-to-cell transmission in a T-cell model. Thus, our work demonstrates that interactions involving Tax-1 PDZ-domains are amenable to small-molecule inhibition, which provides a framework for the design of targeted therapies for HTLV-1-induced diseases.
- Laboratory of Viral Interactomes, Unit of Molecular Biology of Diseases, GIGA Institute, University of Liege, Liège, Belgium.
Organizational Affiliation: