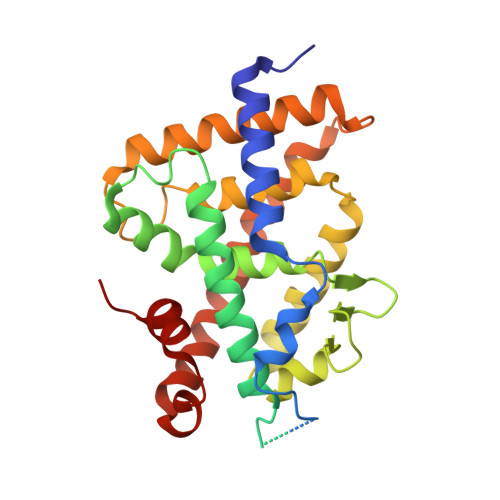
Structural analysis and biological activities of C25-amino and C25-nitro vitamin D analogs.
Gomez-Bouzo, U., Belorusova, A.Y., Rivadulla, M.L., Santalla, H., Verlinden, L., Verstuyf, A., Ferronato, M.J., Curino, A.C., Facchinetti, M.M., Fall, Y., Gomez, G., Rochel, N.(2023) Bioorg Chem 136: 106528-106528
- PubMed: 37054528
- DOI: https://doi.org/10.1016/j.bioorg.2023.106528
- Primary Citation of Related Structures:
8CK5, 8CKC - PubMed Abstract:
Intense synthetic efforts have been directed towards the development of noncalcemic analogs of 1,25-dihydroxyvitamin D 3 . We describe here the structural analysis and biological evaluation of two derivatives of 1,25-dihydroxyvitamin D 3 with modifications limited to the replacement of the 25-hydroxyl group by a 25-amino or 25-nitro groups. Both compounds are agonists of the vitamin D receptor. They mediate biological effects similar to 1,25-dihydroxyvitamin D 3 , the 25-amino derivative being the most potent one while being less calcemic than 1,25-dihydroxyvitamin D 3 . The in vivo properties of the compounds make them of potential therapeutic value.
- Departamento de Química Orgánica and Instituto de Investigación Sanitaria Galicia Sur (IISGS), Campus Lagoas Marcosende, Universidad de Vigo, 36310 Vigo, Spain.
Organizational Affiliation: